Search Count: 16
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2023-03-29 Classification: TRANSFERASE Ligands: SO4, PMP |
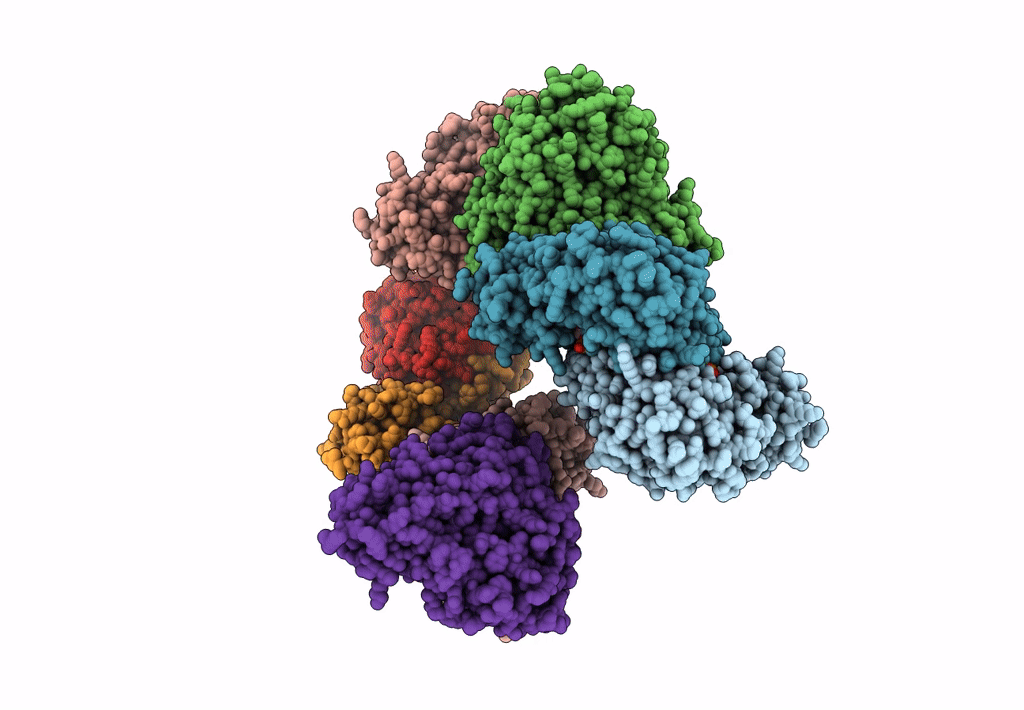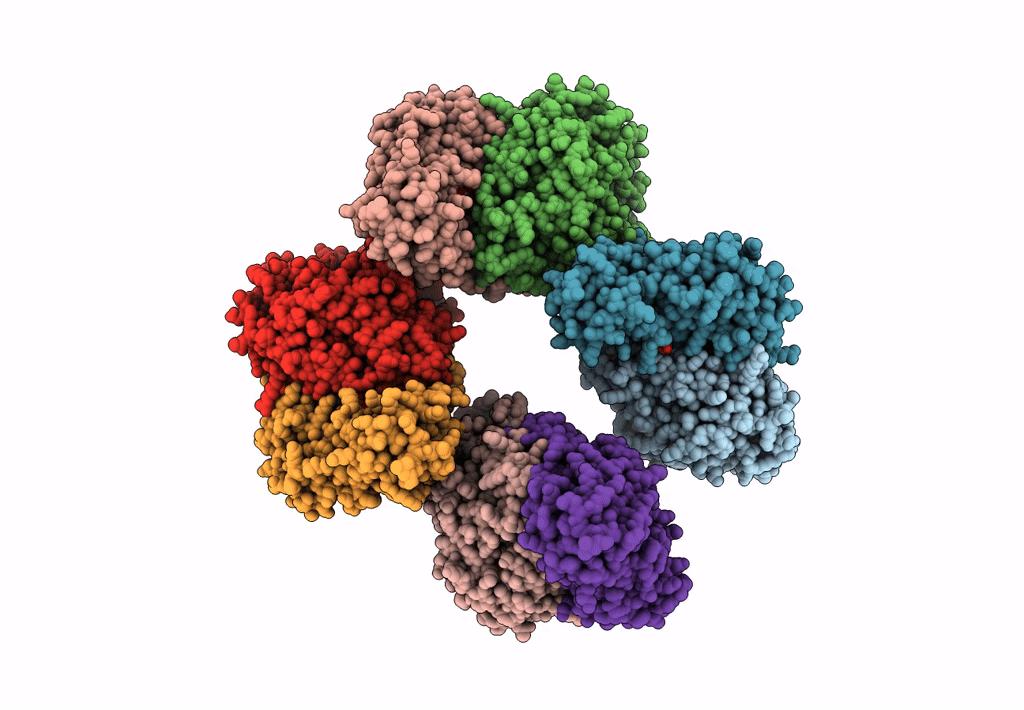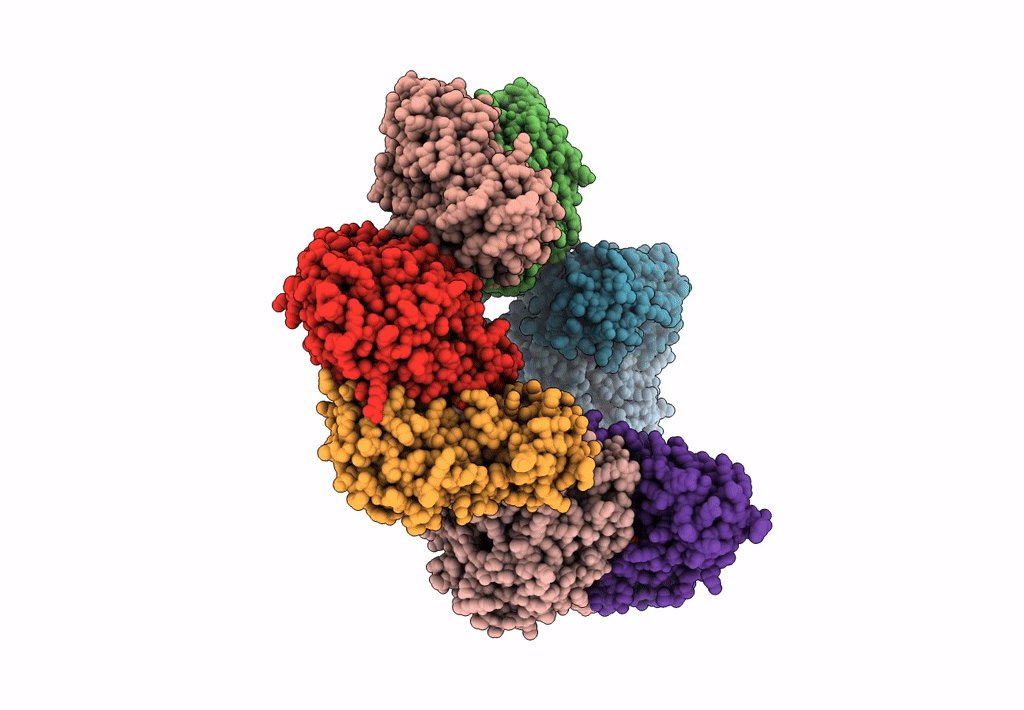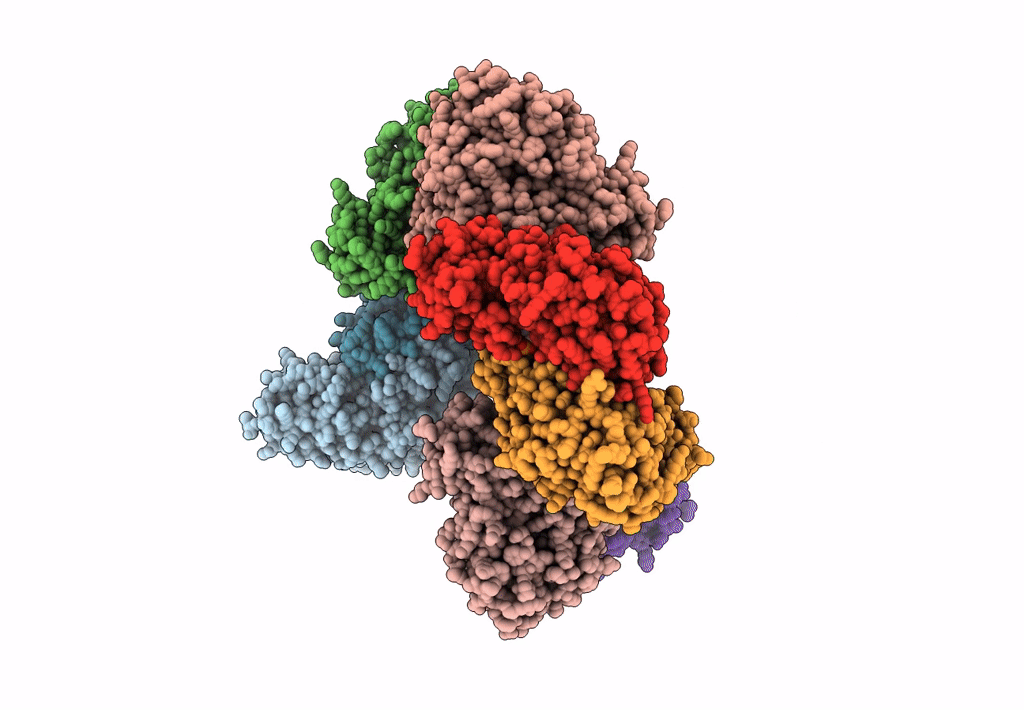 |
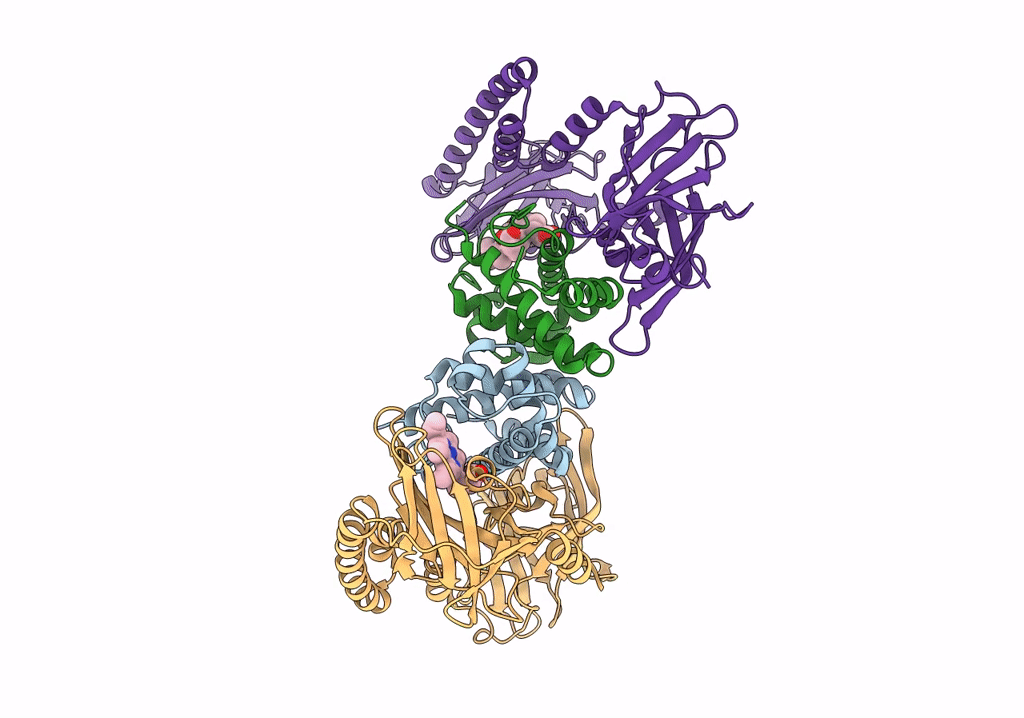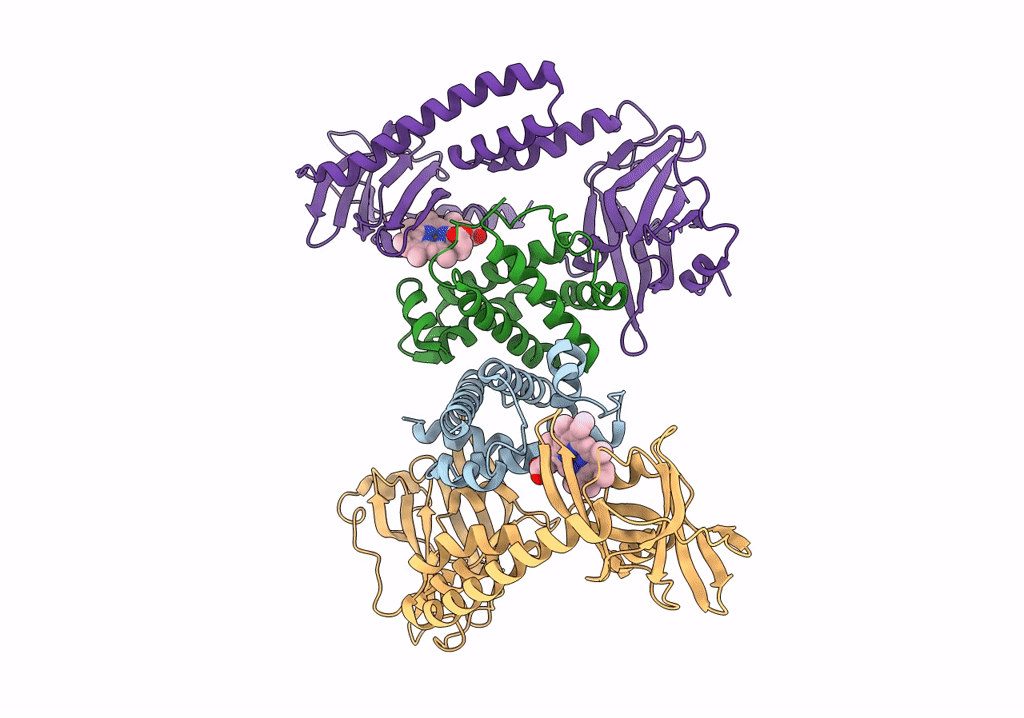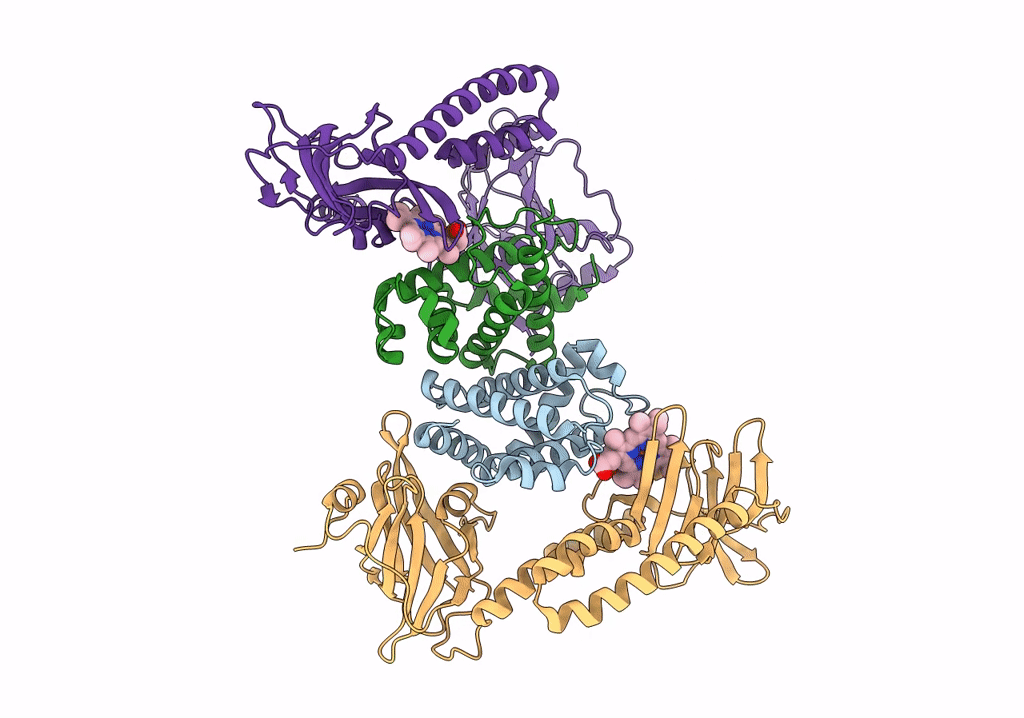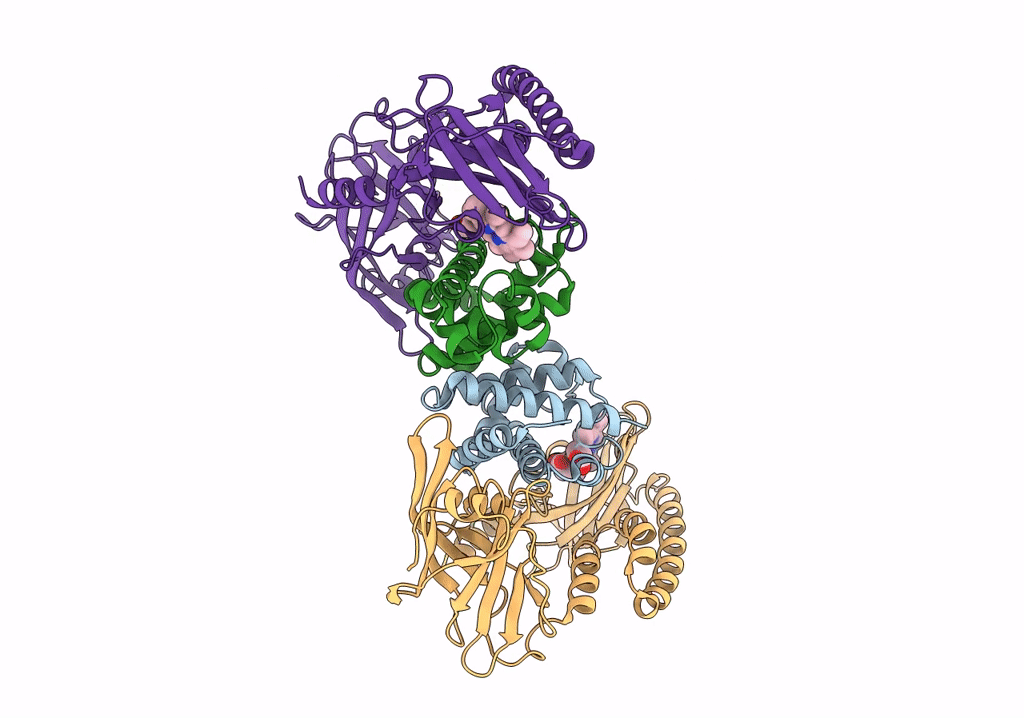
Crystal Structure Of The Human Phosphoserine Aminotransferase (Psat) In Complex With O-Phosphoserine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2023-03-29 Classification: TRANSFERASE Ligands: E1U, SEP, PMP, SO4 |
 |
Organism: Staphylococcus aureus subsp. aureus mw2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-04-13 Classification: METAL TRANSPORT Ligands: HEM |
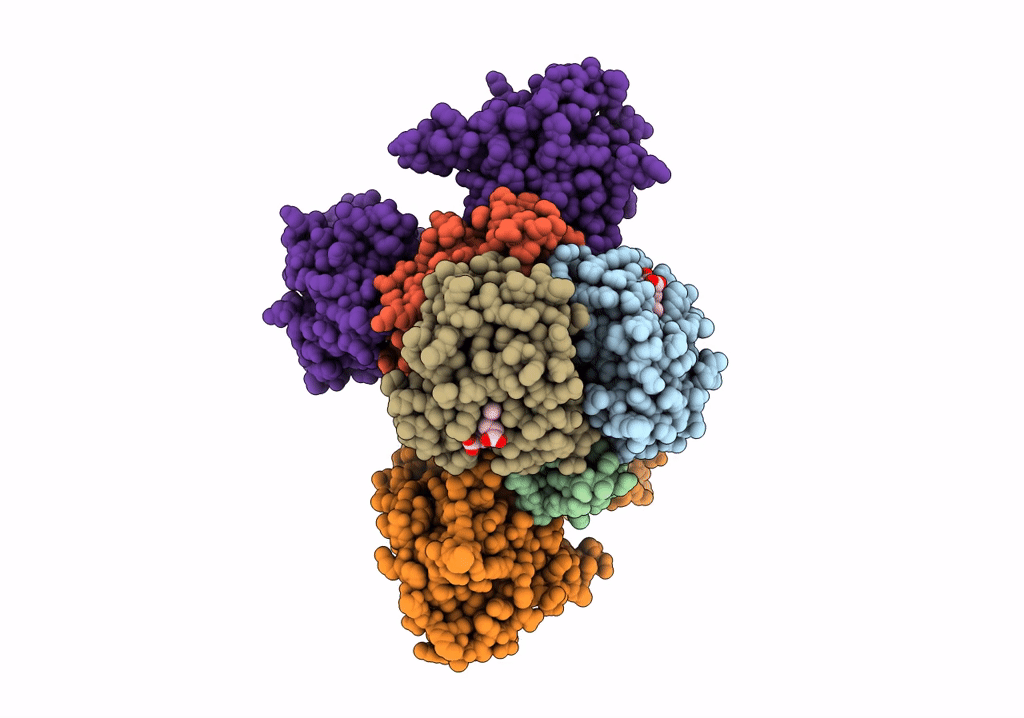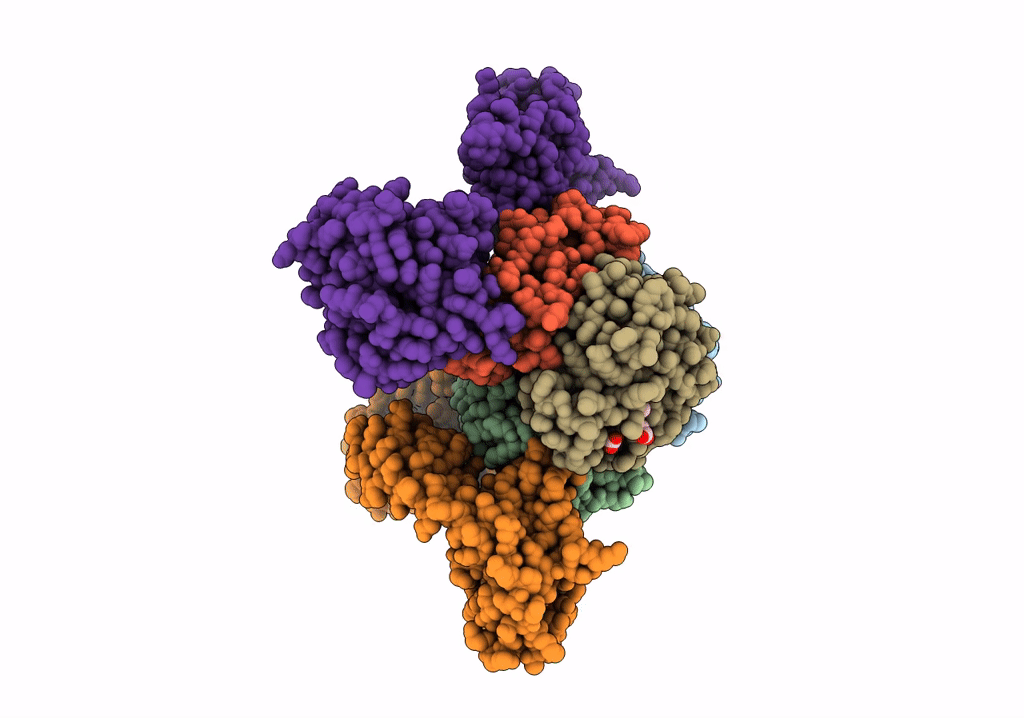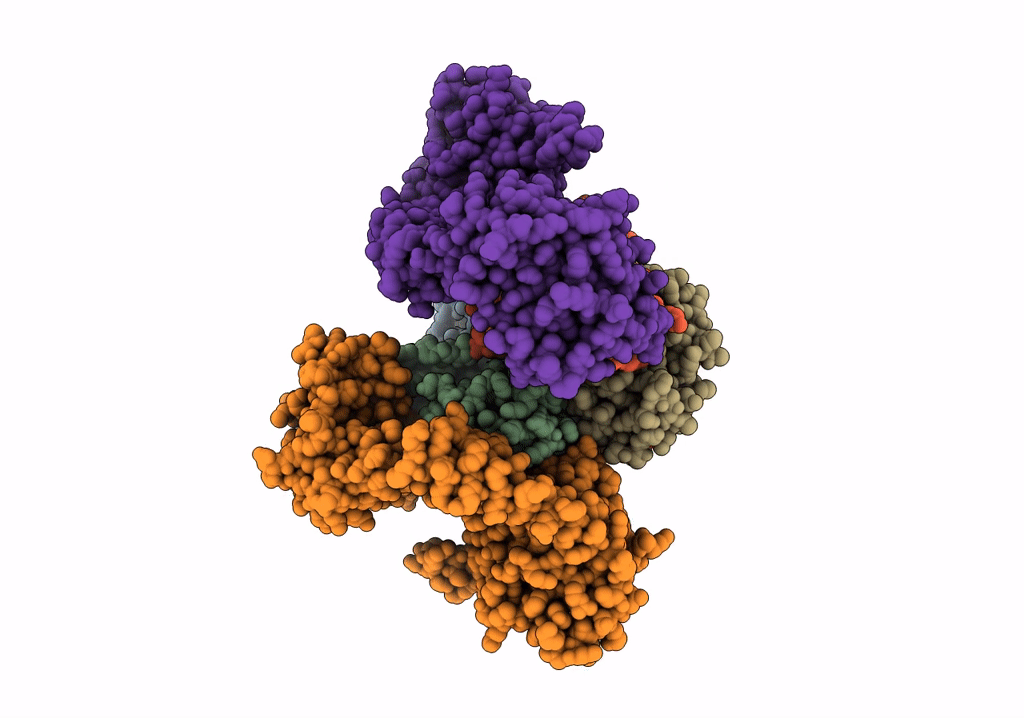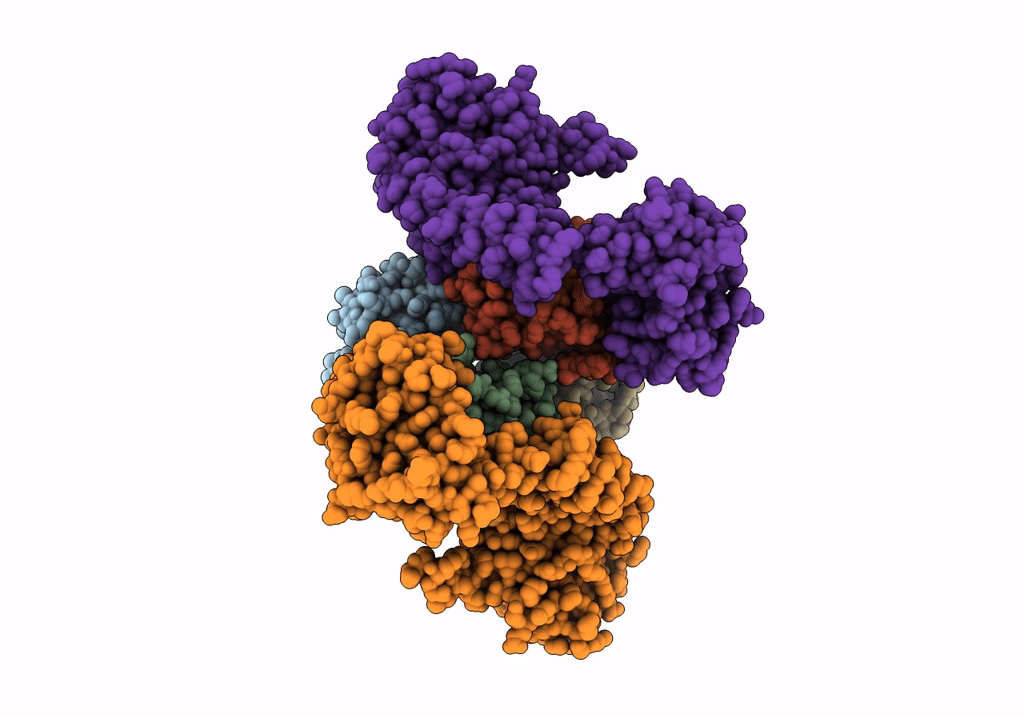 |
Human Carboxyhemoglobin Bound To Staphylococcus Aureus Hemophore Isdb - 1:2 Complex
Organism: Staphylococcus aureus subsp. aureus mw2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-04-13 Classification: METAL TRANSPORT Ligands: HEM |
 |
Human Carboxyhemoglobin Bound To Staphylococcus Aureus Hemophore Isdb - 1:1 Complex
Organism: Staphylococcus aureus subsp. aureus mw2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-04-13 Classification: METAL TRANSPORT Ligands: HEM |
 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium str. lt2
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2021-04-07 Classification: LYASE Ligands: PLP, Q7B, DMS, GOL, CO |
 |
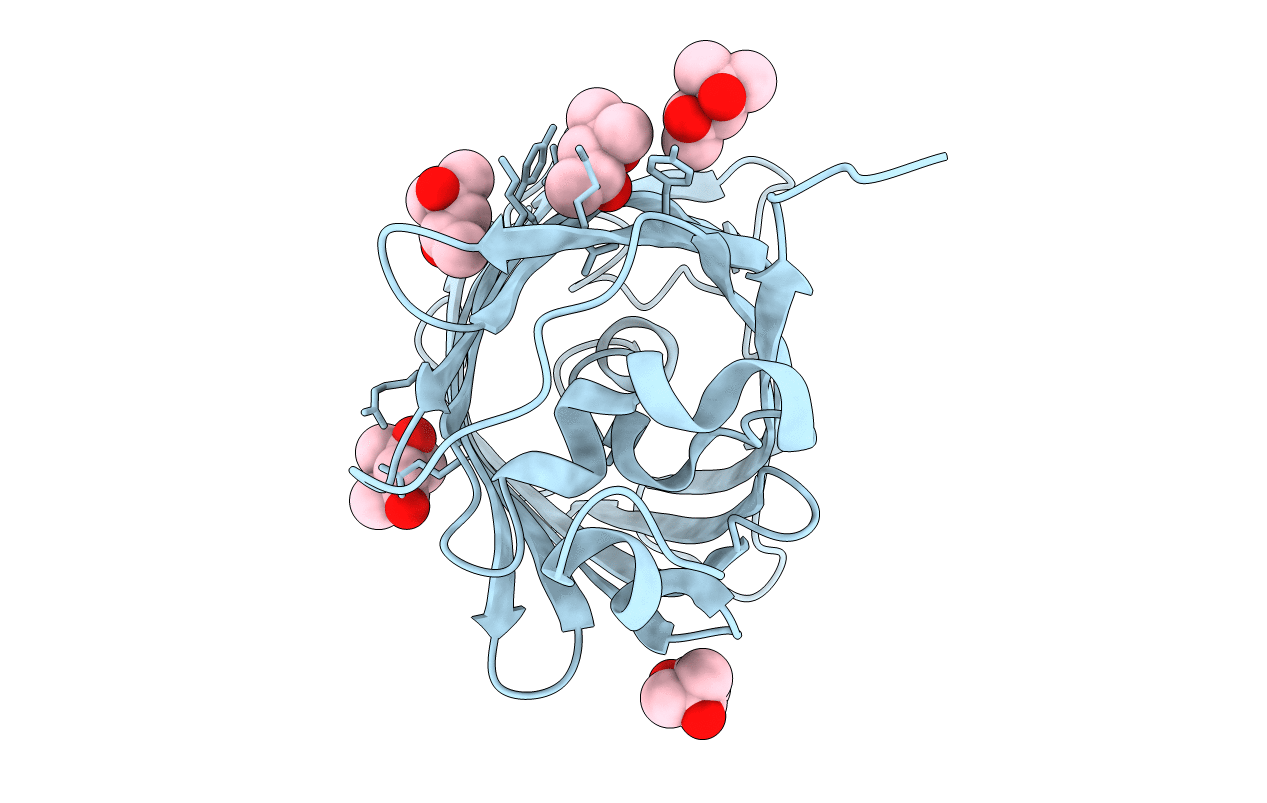
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2018-12-19 Classification: FLUORESCENT PROTEIN Ligands: MPD, MRD |
 |
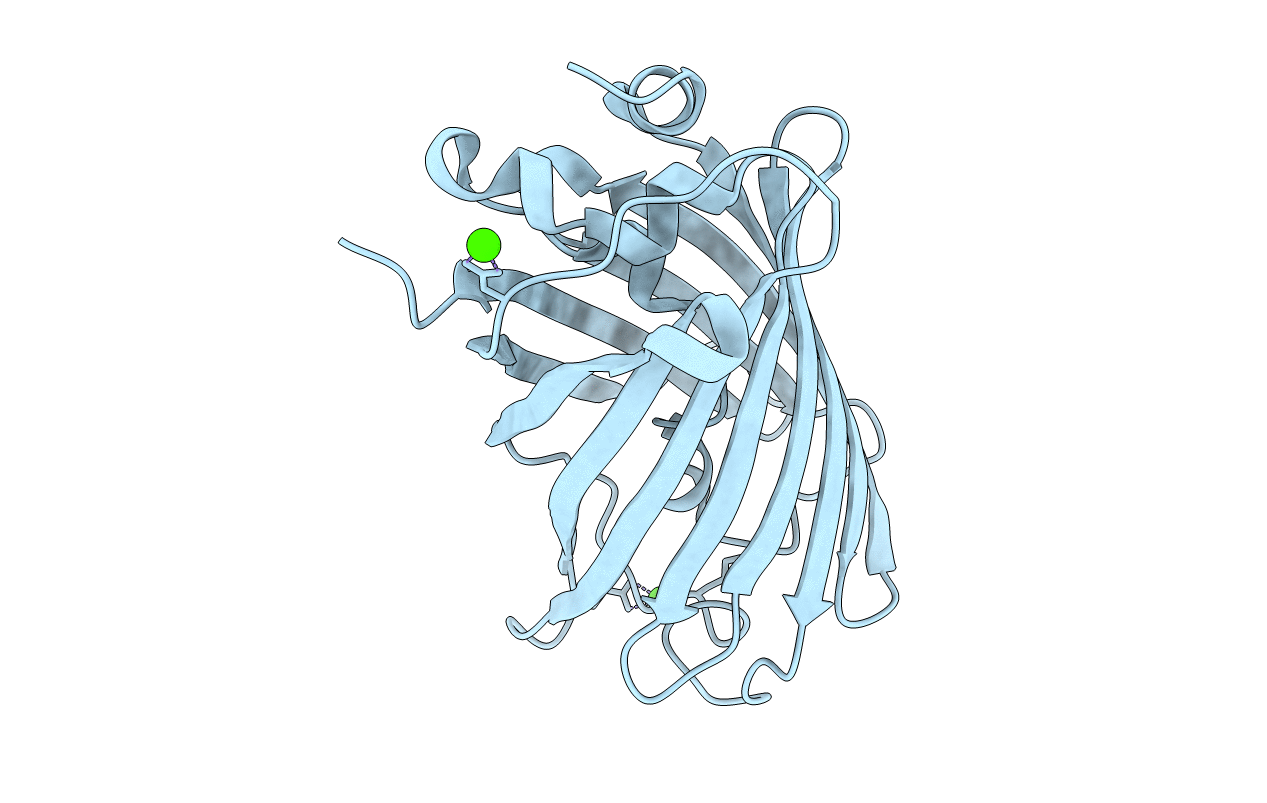
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2018-12-19 Classification: FLUORESCENT PROTEIN Ligands: MPD, MRD |
 |
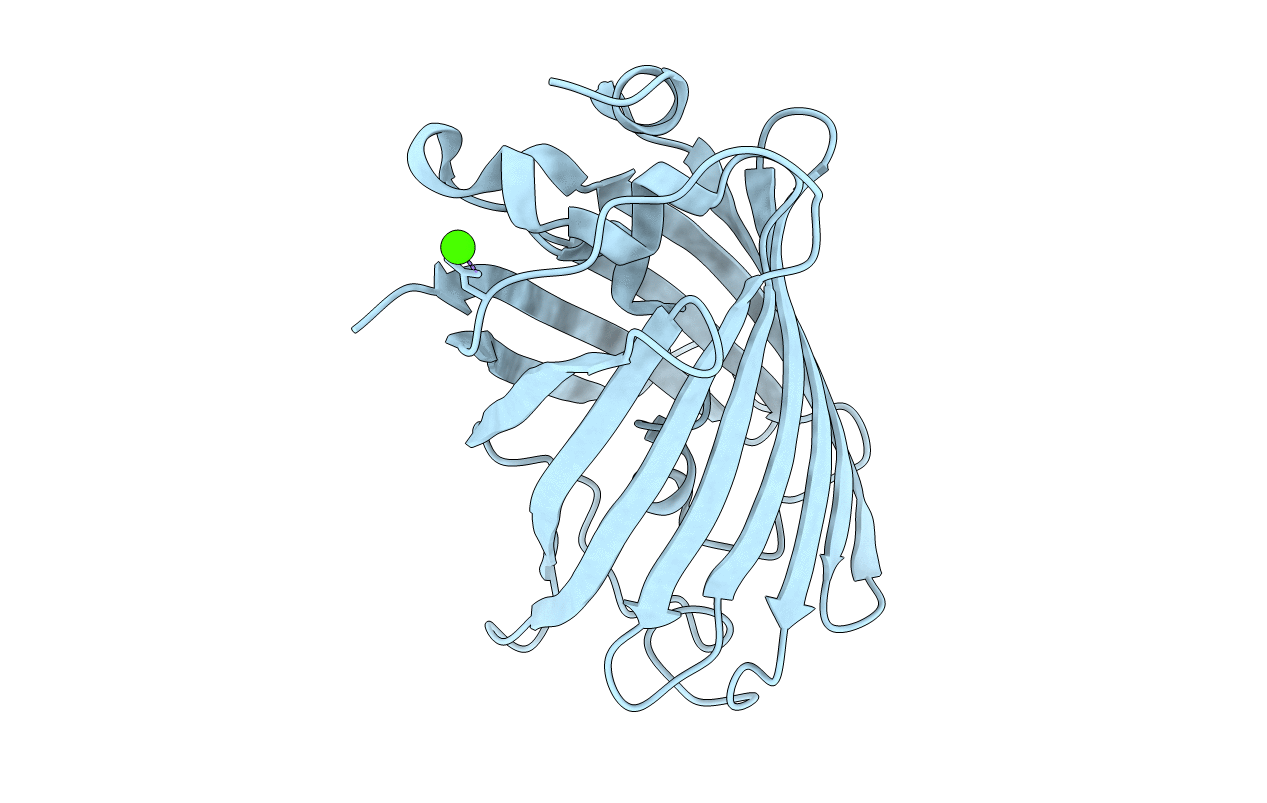
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2018-12-19 Classification: FLUORESCENT PROTEIN Ligands: CA |
 |
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2018-12-19 Classification: FLUORESCENT PROTEIN Ligands: CA |
 |
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-12-19 Classification: FLUORESCENT PROTEIN |
 |
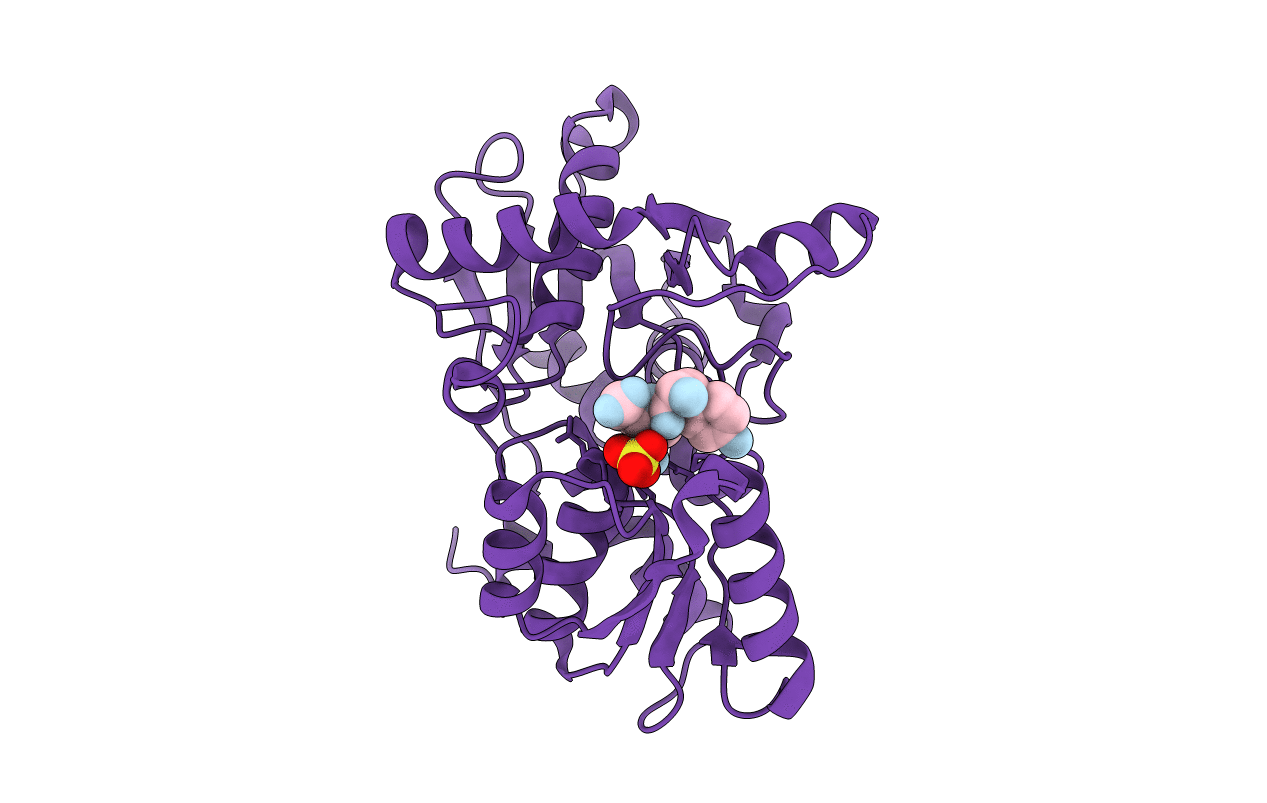
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2009-11-17 Classification: TRANSFERASE |
 |
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2009-11-17 Classification: TRANSFERASE Ligands: SO4 |
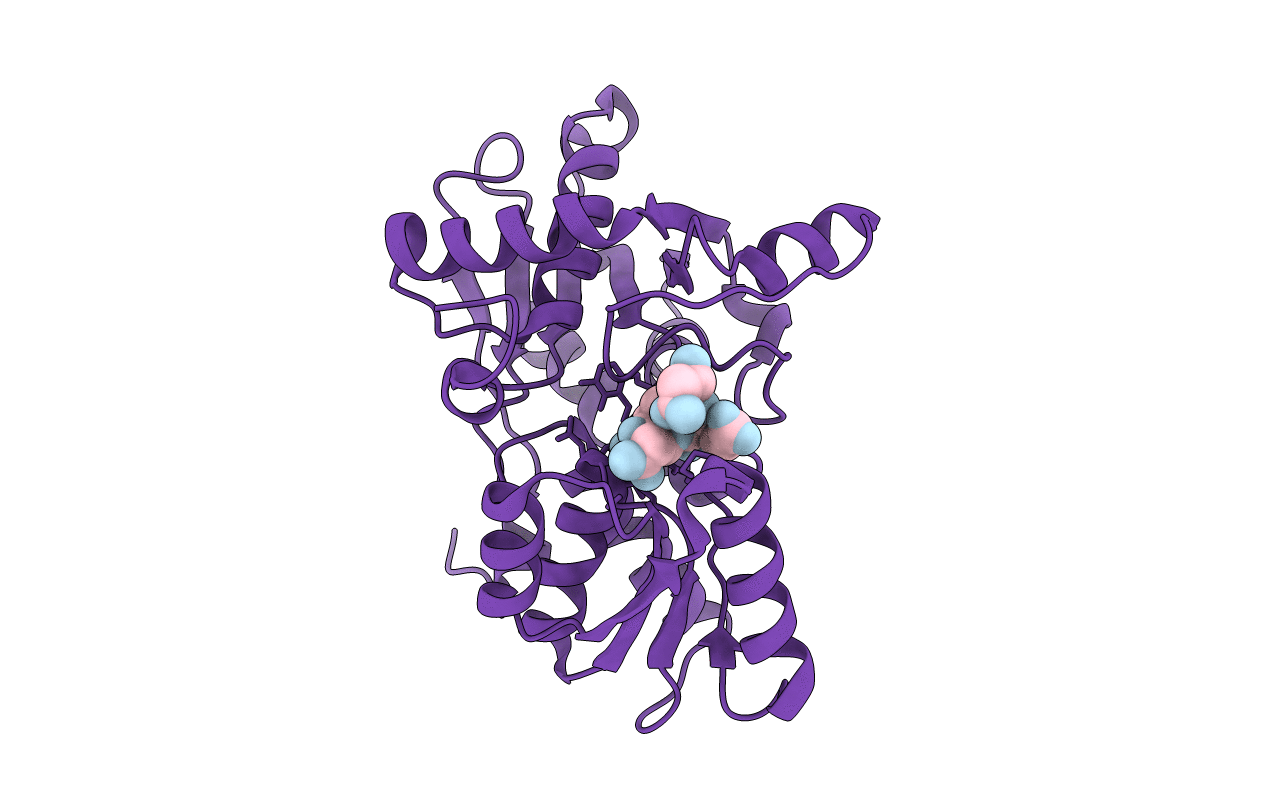 |
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2009-11-17 Classification: TRANSFERASE |
 |
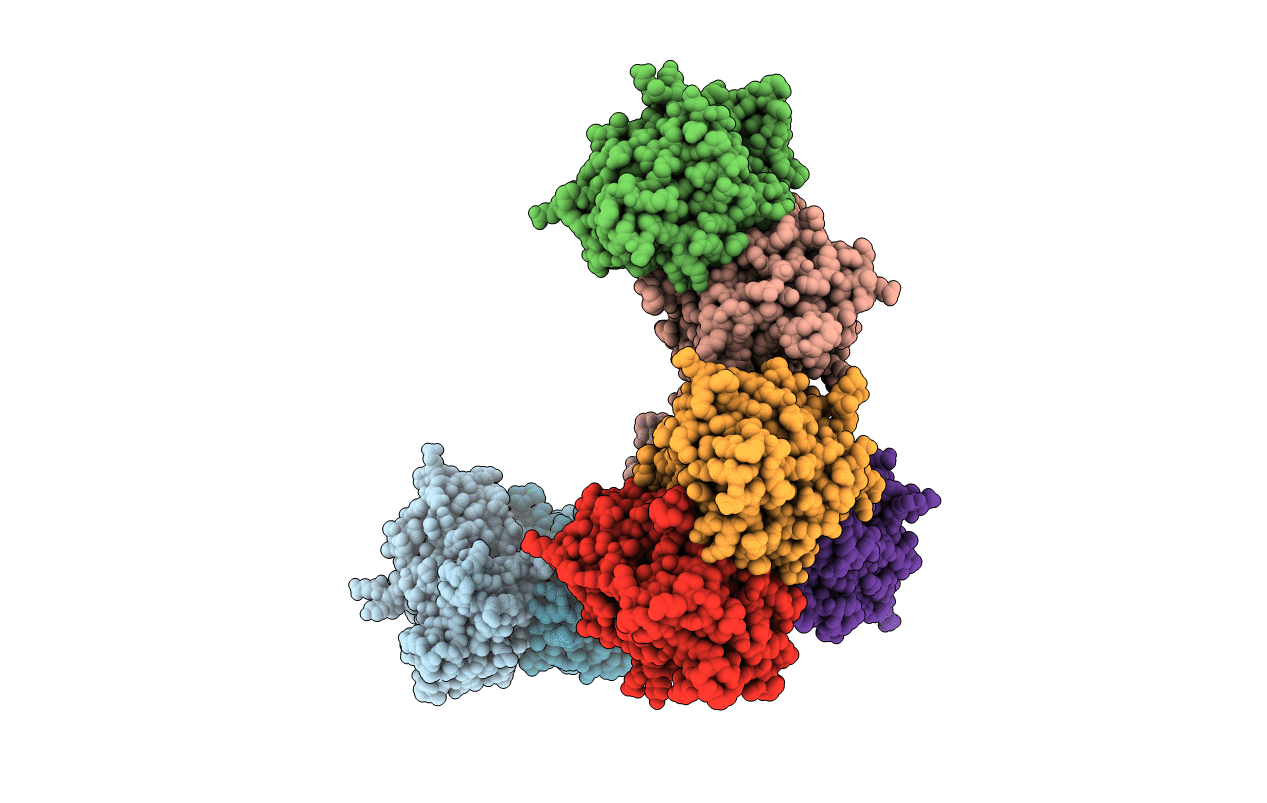
Green Fluorescent Protein Ground States: The Influence Of A Second Protonation Site Near The Chromophore
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2007-04-24 Classification: LUMINESCENT PROTEIN Ligands: SO4 |
 |
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2007-01-23 Classification: TRANSFERASE Ligands: PLP |

