Search Count: 78
 |
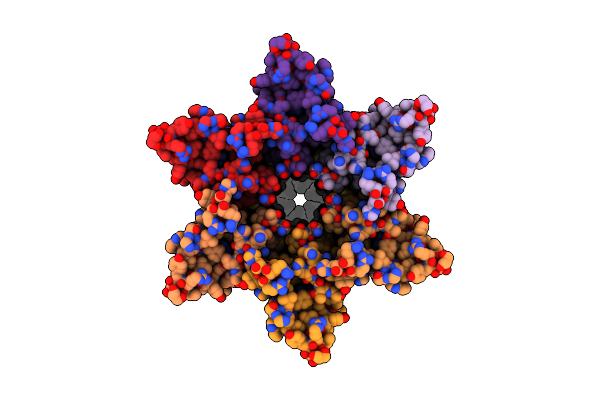
Cryo-Em Structure Of Cx26 Gap Junction K125E Mutant In Bicarbonate Buffer (Classification On Hemichannel)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-12 Classification: MEMBRANE PROTEIN Ligands: LMT, PTY |
 |
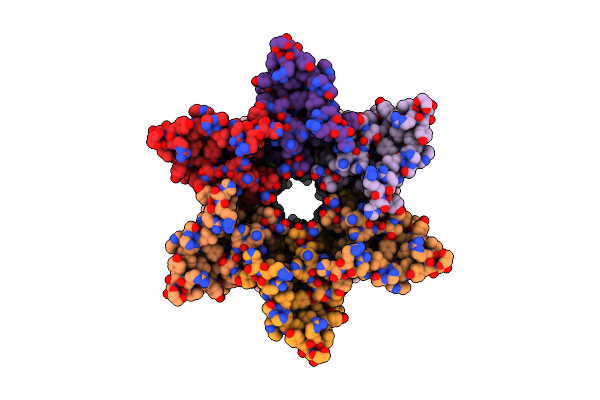
Cryo-Em Structure Of Cx26 Solubilised In Lmng - Hemichannel Classification - Nconst Conformation
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-12 Classification: MEMBRANE PROTEIN Ligands: PTY |
 |
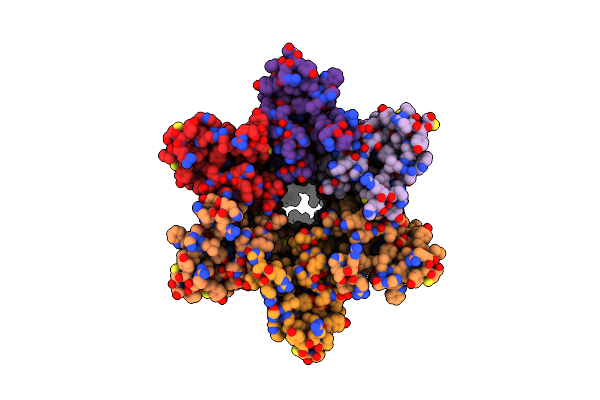
Cryo-Em Structure Of Cx26 Solubilised In Lmng - Hemichannel Classification Nflex Conformation
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-12 Classification: MEMBRANE PROTEIN Ligands: PTY |
 |
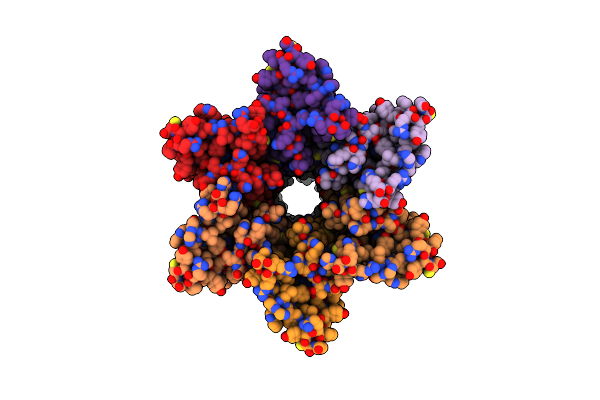
Cryo-Em Structure Of Cx26 Solubilised In Lmng: Classification On Subunit A; Nconst-Mon Conformation
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-12 Classification: MEMBRANE PROTEIN Ligands: PTY |
 |
Cryo-Em Structure Of Cx26 Solubilised In Lmng: Classification On Subunit A; Nflex Conformation
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-12 Classification: MEMBRANE PROTEIN Ligands: PTY |
 |
Organism: Neisseria meningitidis mc58
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-07-12 Classification: TRANSPORT PROTEIN |
 |
Organism: Neisseria meningitidis mc58
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-07-12 Classification: TRANSPORT PROTEIN |
 |
Structure Of Codb, A Cytosine Transporter In An Outward-Facing Conformation
Organism: Proteus vulgaris
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-07-13 Classification: TRANSPORT PROTEIN Ligands: CYT, PEF, LMT, A6L, NA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-06-22 Classification: MEMBRANE PROTEIN Ligands: LMT, PTY |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-06-15 Classification: MEMBRANE PROTEIN Ligands: LMT, PTY |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-06-15 Classification: MEMBRANE PROTEIN Ligands: LMT, PTY |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-06-15 Classification: MEMBRANE PROTEIN Ligands: LMT, PTY |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-03-30 Classification: MEMBRANE PROTEIN Ligands: LMT, PTY |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-03-30 Classification: MEMBRANE PROTEIN Ligands: LMT, PTY |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-03-30 Classification: MEMBRANE PROTEIN Ligands: LMT, PTY |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-03-30 Classification: MEMBRANE PROTEIN Ligands: LMT, PTY |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-03-30 Classification: MEMBRANE PROTEIN Ligands: LMT, PTY |
 |
Crystal Structure Of Lpqy From Mycobacterium Thermoresistible In Complex With Trehalose
Organism: Mycolicibacterium thermoresistibile
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-04-28 Classification: TRANSPORT PROTEIN |
 |
The Structure Of A Quaternary Ammonium Rieske Monooxygenase Reveals Insights Into Carnitine Oxidation By Gut Microbiota And Inter-Subunit Electron Transfer
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-03-31 Classification: OXIDOREDUCTASE Ligands: FE, 152, SCN, FES |
 |
Crystal Structure Of The Apo Form Of A Quaternary Ammonium Rieske Monooxygenase Cnta
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2020-11-18 Classification: OXIDOREDUCTASE Ligands: FES |

