Search Count: 63
 |
Organism: Myxococcus xanthus dz2
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2026-01-28 Classification: TRANSLOCASE Ligands: GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2025-07-30 Classification: HYDROLASE Ligands: CL, ZN, MES, EDO, PEG, MPD |
 |
Crystal Structure Of Human Ribonuclease 7 (Rnase 7) In Complex With 5'-Adenosine Monophosphate (5'-Amp)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2025-07-30 Classification: HYDROLASE Ligands: BO3, AMP, GOL, EDO |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-06-26 Classification: MEMBRANE PROTEIN Ligands: C8E |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:3.96 Å Release Date: 2024-06-26 Classification: MEMBRANE PROTEIN |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2024-06-26 Classification: MEMBRANE PROTEIN Ligands: C8E, MG |
 |
C2221 Crystal Structure Of Tama (Barrel Only) From Pseudomonas Aeruginosa At 3.15 Ang
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2024-06-26 Classification: MEMBRANE PROTEIN Ligands: PT |
 |
Crystal Structure Of A Resurrected Ancestor (Ancrnase) Of The Pancreatic-Type Rnases 2 And 3 Sub-Families
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2024-02-28 Classification: HYDROLASE Ligands: SO4, ACT, MPD |
 |
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2024-01-31 Classification: CELL ADHESION Ligands: ACT, GOL |
 |
Crystal Structure Of Human Eosinophil-Derived Neurotoxin (Edn, Ribonuclease 2) In Complex With 5'-Adenosine Monophosphate (Amp)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-11-29 Classification: HYDROLASE Ligands: SIN, EDO, AMP |
 |
Crystal Structure Of Apo Eosinophil Cationic Protein (Ribonuclease 3) From Macaca Fascicularis (Mfecp)
Organism: Macaca fascicularis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-08-16 Classification: HYDROLASE Ligands: GOL, CIT, PEG |
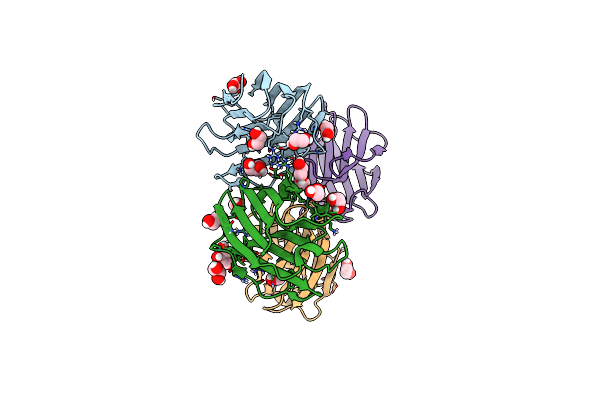 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-02-01 Classification: SUGAR BINDING PROTEIN Ligands: GOL, EDO |
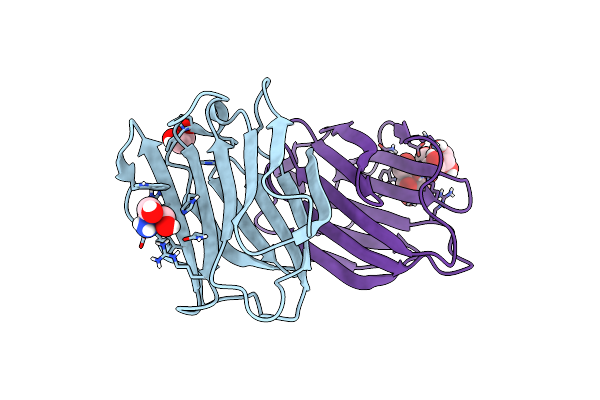 |
Crystal Structure Of R14A-R20A Human Galectin-7 Mutant In Presence Of Lactose
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-02-01 Classification: SUGAR BINDING PROTEIN Ligands: EDO, TRS |
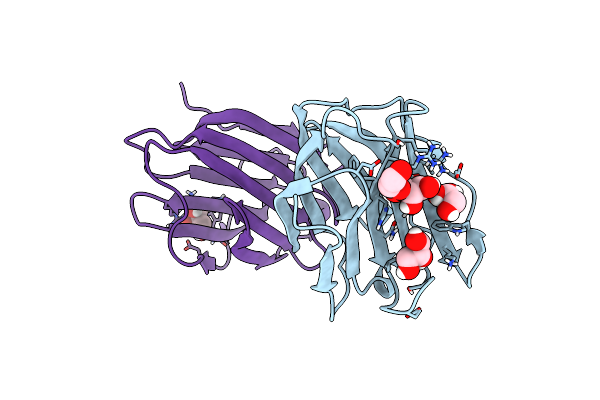 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2023-01-25 Classification: SUGAR BINDING PROTEIN Ligands: EDO, GOL |
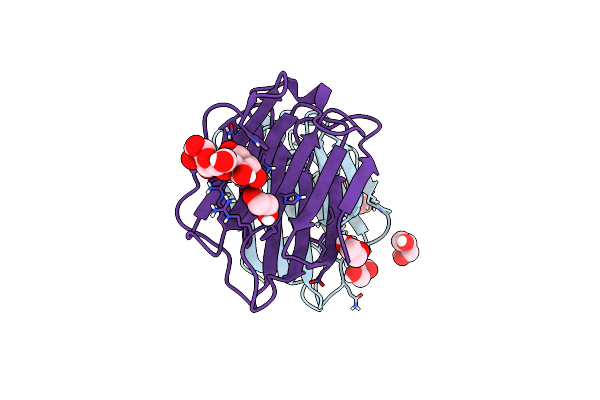 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2023-01-25 Classification: SUGAR BINDING PROTEIN Ligands: GOL, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-01-25 Classification: SUGAR BINDING PROTEIN Ligands: GOL, TRS, EDO, PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2023-01-25 Classification: SUGAR BINDING PROTEIN Ligands: EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-01-25 Classification: SUGAR BINDING PROTEIN Ligands: EDO, TRS, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2023-01-25 Classification: SUGAR BINDING PROTEIN Ligands: GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2023-01-25 Classification: SUGAR BINDING PROTEIN Ligands: EDO |

