Search Count: 5
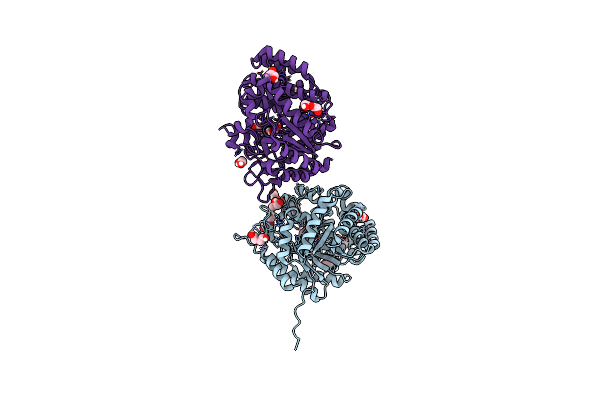 |
The Crystal Structure Of Beta-Glucosidase From The Thermophilic Bacterium Caldicellulosiruptor Saccharolyticus Determined At 1.47 A Resolution
Organism: Caldicellulosiruptor saccharolyticus dsm 8903
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2024-10-16 Classification: HYDROLASE Ligands: GOL, EDO, PGE, 1PE, PEG, CL |
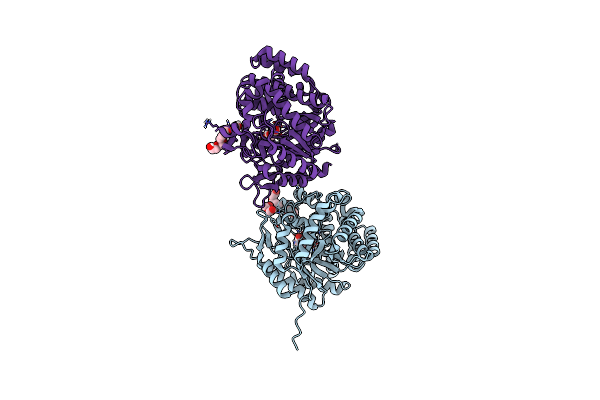 |
The Crystal Structure Of Beta-Glucosidase From The Thermophilic Bacterium Caldicellulosiruptor Saccharolyticus In Complex With Beta-D-Glucose Determined At 1.95 A Resolution
Organism: Caldicellulosiruptor saccharolyticus dsm 8903
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-10-16 Classification: HYDROLASE Ligands: BGC, 1PE, PEG, CL |
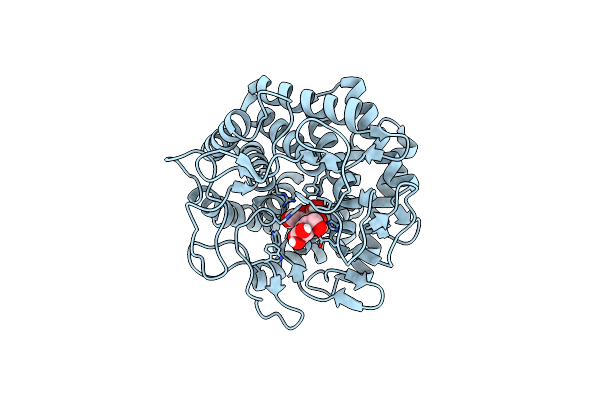 |
Crystal Structure Of The Csce With Ligand To Have A Insight Into The Catalytic Mechanism
Organism: Caldicellulosiruptor saccharolyticus dsm 8903
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2021-12-01 Classification: PROTEIN BINDING Ligands: CL |
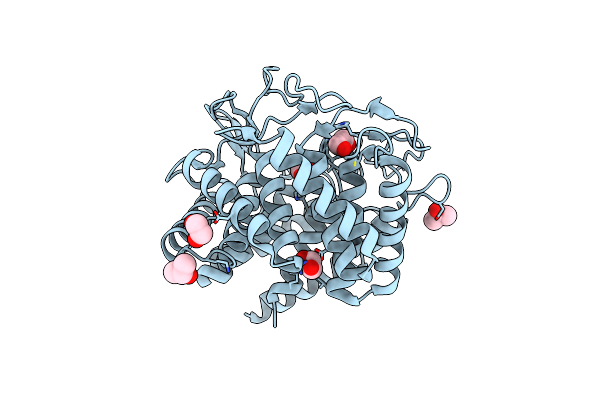 |
Crystal Structure Of Cellobiose 2-Epimerase From Caldicellulosiruptor Saccharolyticus Dsm 8903
Organism: Caldicellulosiruptor saccharolyticus dsm 8903
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2016-04-13 Classification: ISOMERASE Ligands: IPA, EDO |
 |
Crystal Structure Of Cellobiose 2-Epimerase From Caldicellulosiruptor Saccharolyticus Dsm 8903
Organism: Caldicellulosiruptor saccharolyticus dsm 8903
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2016-04-13 Classification: ISOMERASE Ligands: EDO |

