Search Count: 114
 |
Cryo-Em Structure Of Adriforant-Bound Histamine Receptor 4 H4R At Inactive State
Organism: Homo sapiens, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: A1EBW |
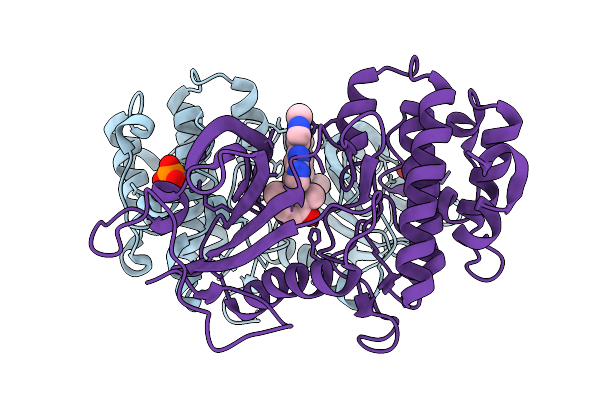 |
Discovery Of A Bifunctional Pkmyt1-Targeting Protac Empowered By Ai-Generation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: PROTEIN BINDING Ligands: A1EMX, PO4, CL |
 |
Cryo-Em Structure Of Histamine-Bound Histamine Receptor 3 H3R G Protein Complex
Organism: Homo sapiens, Bos taurus, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: MEMBRANE PROTEIN Ligands: HSM |
 |
Cryo-Em Structure Of Histamine-Bound Histamine Receptor 4 H4R G Protein Complex
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: HSM, PO4 |
 |
Organism: Horseshoe bat sarbecovirus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: VIRAL PROTEIN Ligands: NAG, EIC |
 |
Organism: Coronaviridae
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: VIRAL PROTEIN Ligands: NAG |
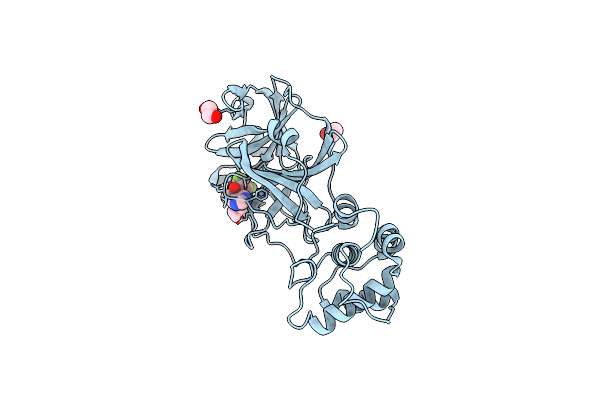 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2025-04-23 Classification: VIRAL PROTEIN Ligands: XDQ, EDO, CL |
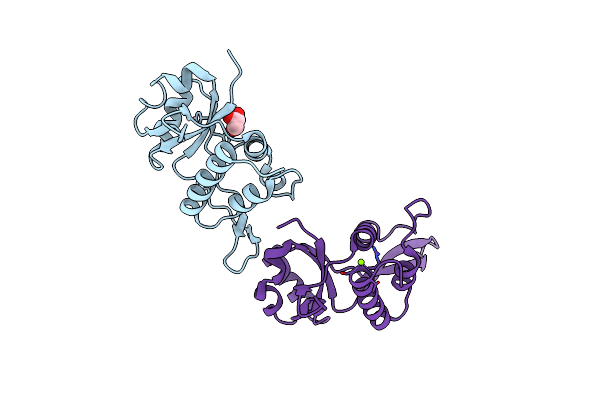 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-03-19 Classification: TRANSCRIPTION Ligands: GOL, MG |
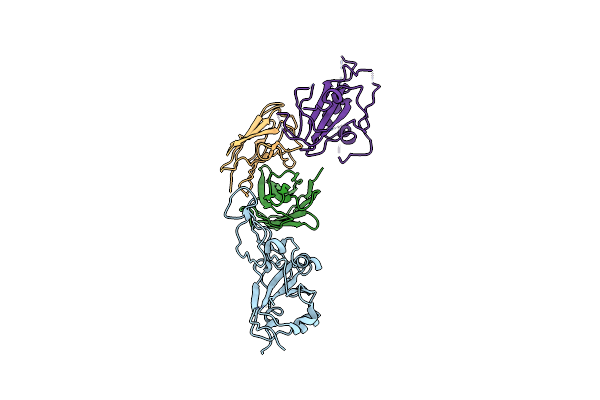 |
Cryo-Em Structure Of The Sars-Cov-2 S 6P Trimer In Complex With The Human Neutralizing Antibody Fab Fragment Cav-C65 (Local Refinement)
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: VIRAL PROTEIN |
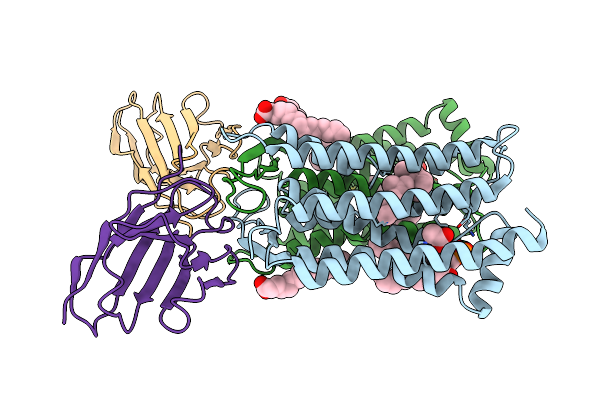 |
Organism: Escherichia coli, Vicugna pacos
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: MEMBRANE PROTEIN Ligands: RET, ACT, PX2, PLM |
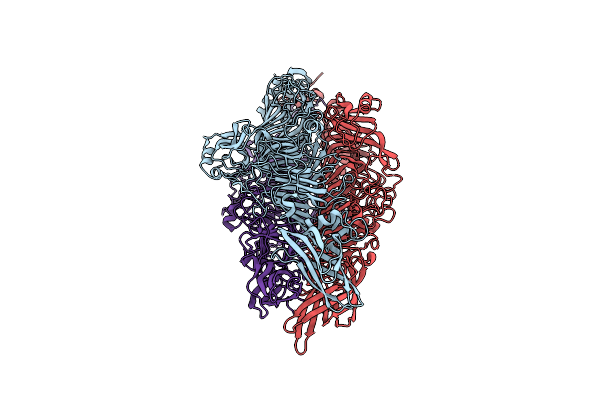 |
Cryo-Em Structure Of A Bacteriophage Tail- Spike Protein Against Klebsiella Pneumoniae K64,Orf41(K64-Orf41)
Organism: Klebsiella phage sh-kp 152410
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: LYASE |
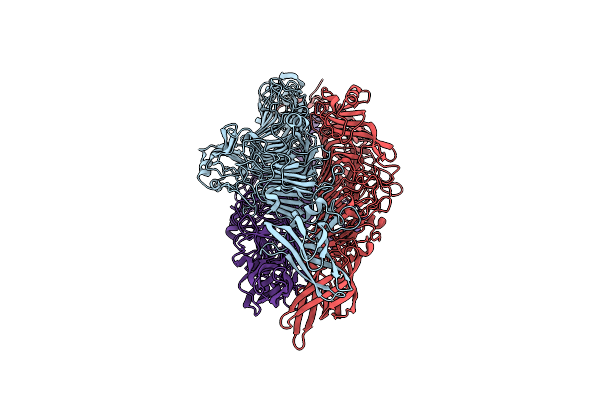 |
Cryo-Em Structure Of A Bacteriophage Tail- Spike Protein Against Klebsiella Pneumoniae K64,Orf41(K64-Orf41) In 5 Mm Edta
Organism: Klebsiella phage sh-kp 152410
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: LYASE |
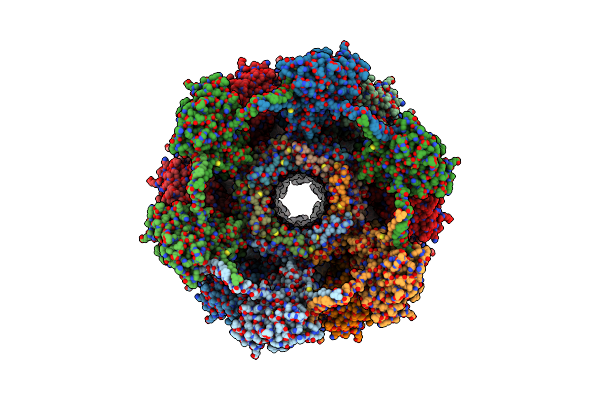 |
Organism: Clostridioides difficile
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRUS LIKE PARTICLE |
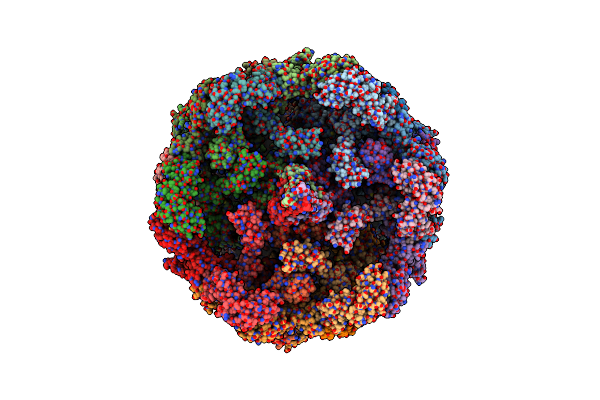 |
Cryoem Structure Of Diffocin - Precontracted - Baseplate - Focused Refinement On Triplex Region
Organism: Clostridioides difficile
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRUS LIKE PARTICLE |
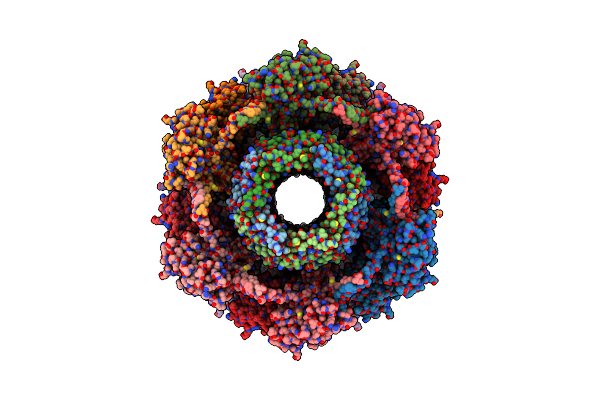 |
Organism: Clostridioides difficile
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRUS LIKE PARTICLE |
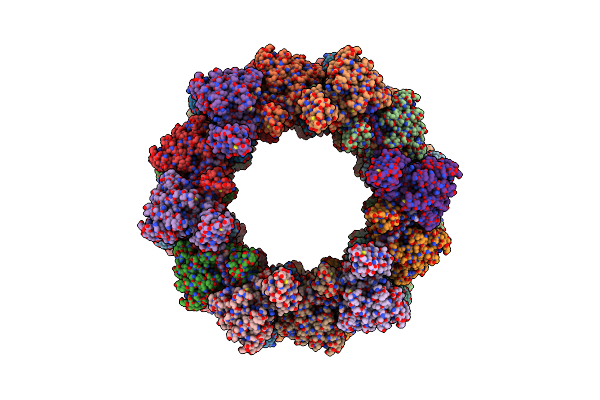 |
Organism: Clostridioides difficile
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRUS LIKE PARTICLE |
 |
Cryoem Structure Of Diffocin - Postcontracted - Collar - Transitional State
Organism: Clostridioides difficile
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRUS LIKE PARTICLE |
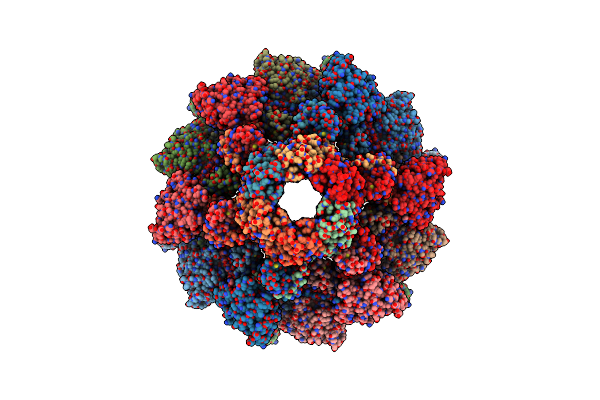 |
Organism: Clostridioides difficile
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRUS LIKE PARTICLE |
 |
Cryoem Structure Of Diffocin - Postcontracted - Baseplate - Transitional State
Organism: Clostridioides difficile
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Clostridioides difficile
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRUS LIKE PARTICLE |

