Search Count: 63
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: BIOSYNTHETIC PROTEIN |
 |
Crystal Structure Of Beta-Ketoacyl-Acp Synthase Fabh C112Q In Complex With Acyl-Acp From E. Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: BIOSYNTHETIC PROTEIN Ligands: 6VG |
 |
Crystal Structure Of Beta-Ketoacyl-Acp Synthase Fabh C112Q In Complex With Malonyl-Acp From E. Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: BIOSYNTHETIC PROTEIN Ligands: A1EE0 |
 |
Organism: Bacillus phage spo1
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: VIRUS |
 |
Crystal Structure Of Dehydrogenase/Isomerase Fabx From Helicobacter Pylori In Complex With Inhibitor 1991
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: BIOSYNTHETIC PROTEIN Ligands: A1ECY, FMN, SF4, MG |
 |
Crystal Structure Of Dehydrogenase/Isomerase Fabx From Helicobacter Pylori In Complex With Inhibitor 1872
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: BIOSYNTHETIC PROTEIN Ligands: FMN, SF4, A1EEQ |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-03-19 Classification: BIOSYNTHETIC PROTEIN Ligands: IOD |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: IMMUNE SYSTEM Ligands: ZN, NA, CA |
 |
Organism: Escherichia phage fcwl1
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: VIRAL PROTEIN |
 |
Organism: Escherichia phage fcwl1
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: VIRUS |
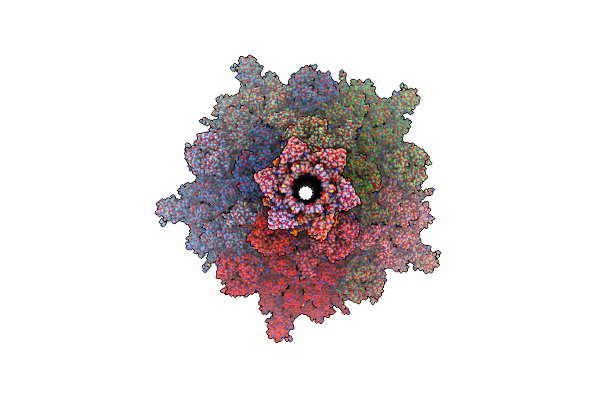 |
The Composite Cryo-Em Structure Of The Portal Vertex Of Bacteriophage Fcwl1
Organism: Escherichia phage fcwl1
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: VIRAL PROTEIN |
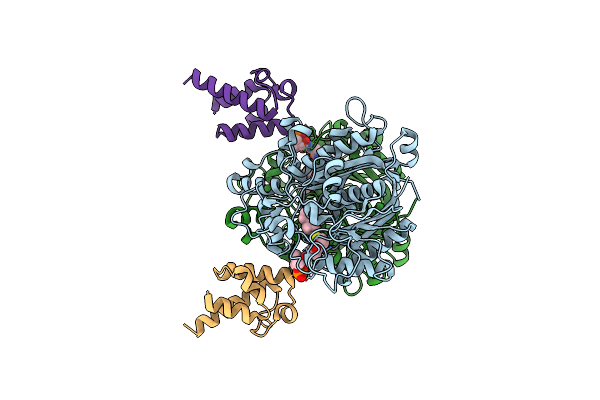 |
Crystal Structure Of Beta-Ketoacyl-Acp Synthase Fabf K336A In Complex With Hexanoyl-Acp From Helicobacter Pylori
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-10-16 Classification: BIOSYNTHETIC PROTEIN Ligands: 6NA, PN7 |
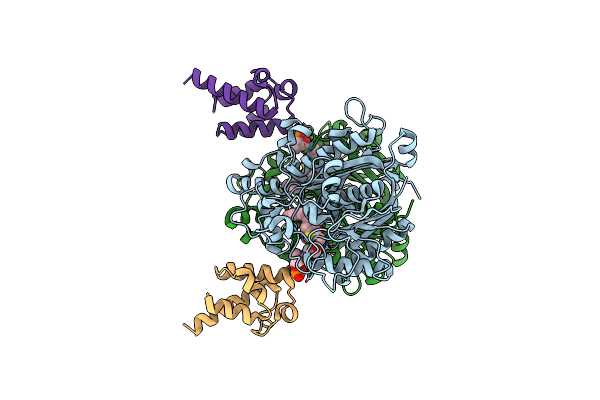 |
Crystal Structure Of Beta-Ketoacyl-Acp Synthase Fabf K336A In Complex With Decanoyl-Acp From Helicobacter Pylori
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-10-16 Classification: BIOSYNTHETIC PROTEIN Ligands: DKA, PN7 |
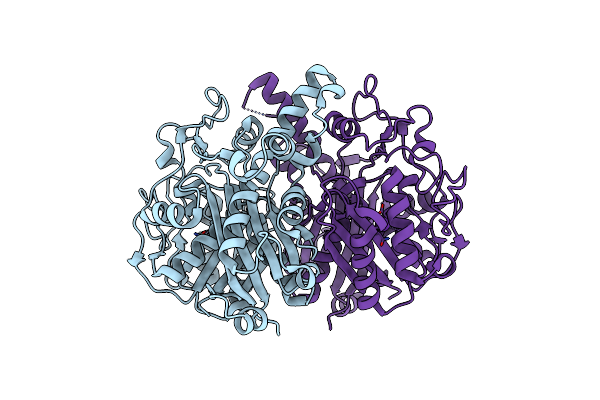 |
Cystal Structure Of Beta-Ketoacyl-Acp Synthase Fabf From Helicobacter Pylori
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-10-09 Classification: BIOSYNTHETIC PROTEIN Ligands: RB |
 |
Crystal Structure Of Beta-Ketoacyl-Acp Synthase Fabf K336A In Complex With Octanoyl-Acp From Helicobacter Pylori
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-10-09 Classification: BIOSYNTHETIC PROTEIN Ligands: OCA, PN7 |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2024-07-10 Classification: ANTIVIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Synthetic construct, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-07-10 Classification: ANTIVIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Deinococcus radiodurans r1 = atcc 13939 = dsm 20539
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-05-15 Classification: HYDROLASE Ligands: SO4, MG |
 |
Crystal Structure Of Deinococcus Radiodurans Exopolyphosphatase Complexed With Pyrophosphate
Organism: Deinococcus radiodurans r1 = atcc 13939 = dsm 20539
Method: X-RAY DIFFRACTION Resolution:3.02 Å Release Date: 2024-05-15 Classification: HYDROLASE Ligands: PPV, MN, SO4, PO4 |

