Search Count: 16
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2024-05-15 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-03-27 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-04-19 Classification: DE NOVO PROTEIN Ligands: EDO |
 |
De Novo Designed Chlorophyll Dimer Protein With Zn Pheophorbide A Methyl Ester, Sp2-Znppam
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-04-19 Classification: DE NOVO PROTEIN Ligands: OE9, PO4, EDO, PGE |
 |
De Novo Designed Chlorophyll Dimer Protein With Zn Pheophorbide A Methyl Ester Matching Geometry Of Purple Bacterial Special Pair, Sp1-Znppam
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-04-19 Classification: DE NOVO PROTEIN Ligands: OE9, SO4, EDO |
 |
Organism: Penicillium aethiopicum
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2016-10-19 Classification: UNKNOWN FUNCTION Ligands: EDO, C8E |
 |
Organism: Penicillium aethiopicum
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2016-10-19 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL, PNS |
 |
Organism: Penicillium aethiopicum
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2016-09-07 Classification: UNKNOWN FUNCTION Ligands: GOL, CL |
 |
Organism: Penicillium aethiopicum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-09-07 Classification: UNKNOWN FUNCTION Ligands: EDO, DMS |
 |
Organism: Aspergillus terreus
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2009-10-27 Classification: TRANSFERASE |
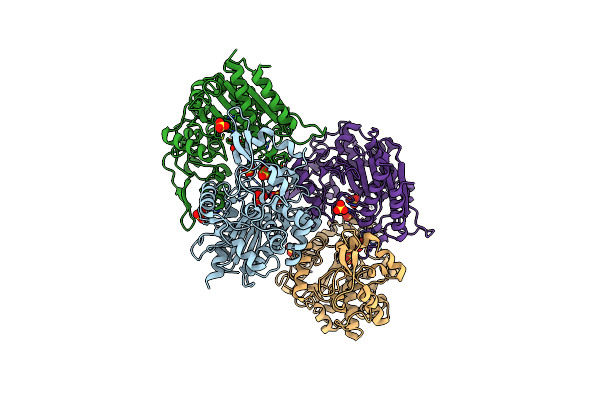 |
Simvastatin Synthase (Lovd) From Aspergillus Terreus, Unliganded, Selenomethionyl Derivative
Organism: Aspergillus terreus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2009-10-27 Classification: TRANSFERASE Ligands: SO4 |
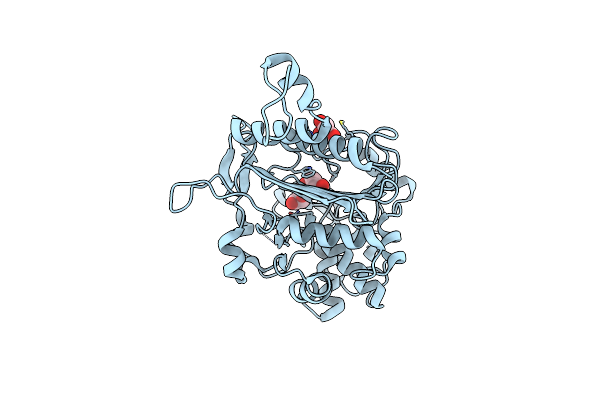 |
Simvastatin Synthase (Lovd) From Aspergillus Terreus, S5 Mutant, Unliganded
Organism: Aspergillus terreus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-10-27 Classification: TRANSFERASE Ligands: GOL, PG4 |
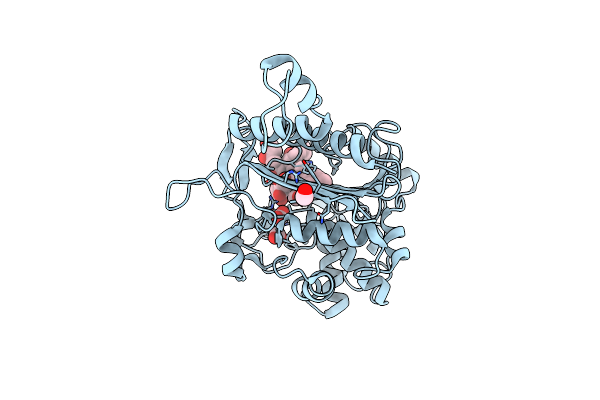 |
Simvastatin Synthase (Lovd), From Aspergillus Terreus, S5 Mutant Complex With Monacolin J Acid
Organism: Aspergillus terreus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-10-27 Classification: TRANSFERASE Ligands: MJA, FMT |
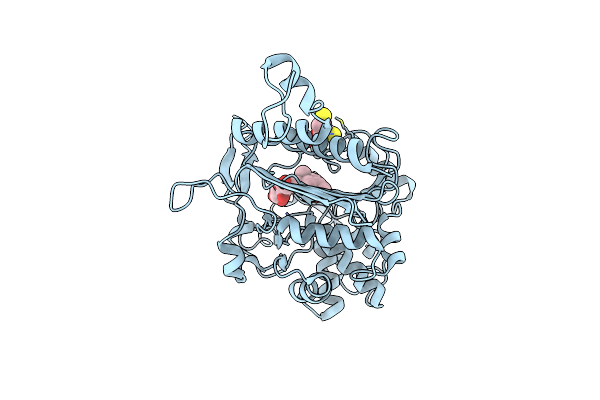 |
Simvastatin Synthase (Lovd), From Aspergillus Terreus, S5 Mutant, S76A Mutant, Complex With Monacolin J Acid
Organism: Aspergillus terreus
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2009-10-27 Classification: TRANSFERASE Ligands: MJA, DTT |
 |
Simvastatin Synthase (Lovd), From Aspergillus Terreus, S5 Mutant, S76A Mutant, Complex With Simvastatin
Organism: Aspergillus terreus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-10-27 Classification: TRANSFERASE Ligands: SIM |
 |
Simvastatin Synthase (Lovd), From Aspergillus Terreus, S5 Mutant, S76A Mutant, Complex With Lovastatin
Organism: Aspergillus terreus
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2009-10-27 Classification: TRANSFERASE Ligands: LVA |

