Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: SIGNALING PROTEIN Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: CYTOKINE Ligands: A1JPQ, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: CYTOKINE Ligands: A1JPS, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: CYTOKINE Ligands: A1JPU |
 |
Organism: Rattus norvegicus, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: CYTOKINE/IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
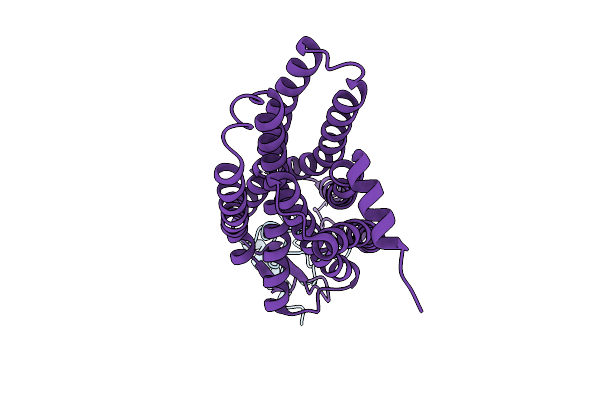 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN |
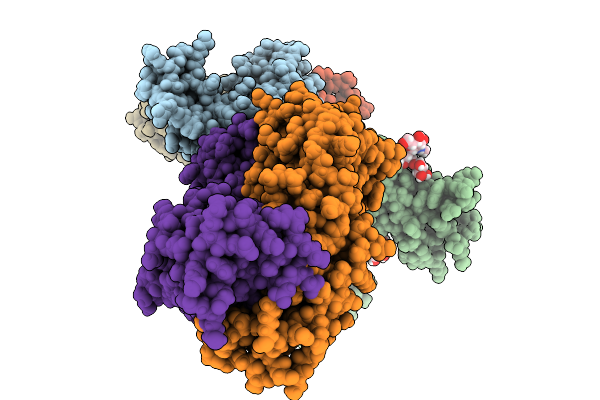 |
Cryo-Em Structure Of The Heterotrimeric Interleukin-2 Receptor In Complex With Interleukin-2 And Anti-Cd25 Fab S417
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: IMMUNE SYSTEM Ligands: NAG |
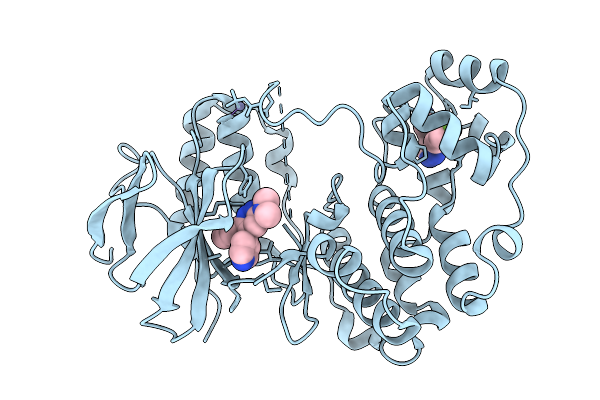 |
Human P38 Alpha Mapk:Mw01-8-105Srm N,N-Dimethyl-6-Phenyl-5-(4-Pyridinyl)-3-Pyridazinamine Inhibitor Complex
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: ZN, GG5, A1CX9 |
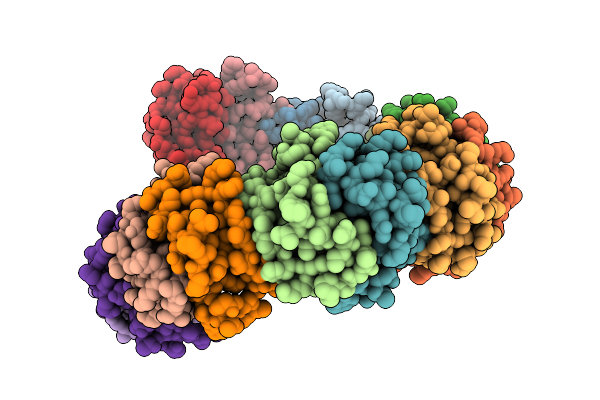 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: CYTOKINE Ligands: NO3 |
 |
 |
Organism: Necator americanus
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: CYTOKINE |
 |
Organism: Homo sapiens, Squalus acanthias
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: Cytokine/Immune System Ligands: EDO, SO4, NI |
 |
Organism: Heligmosomoides polygyrus
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: CYTOKINE |
 |
Organism: Necator americanus
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: ISOMERASE |
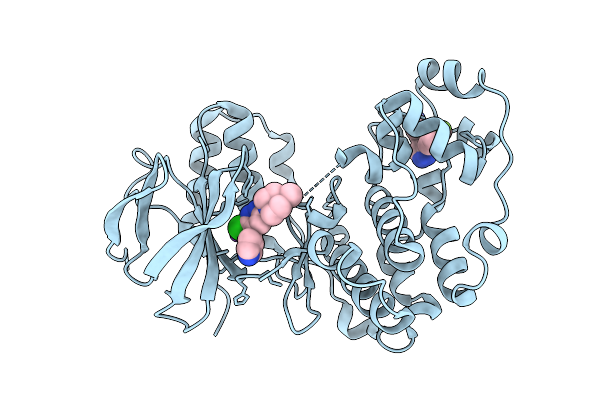 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TRANSFERASE/Inhibitor Ligands: GG5, A1BB6, NA |
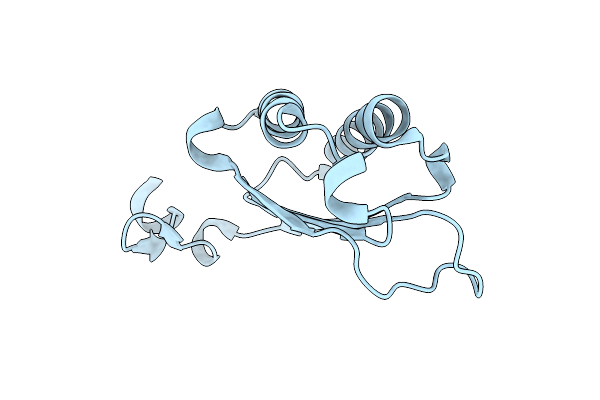 |
Organism: Heligmosomoides polygyrus
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: CYTOKINE |
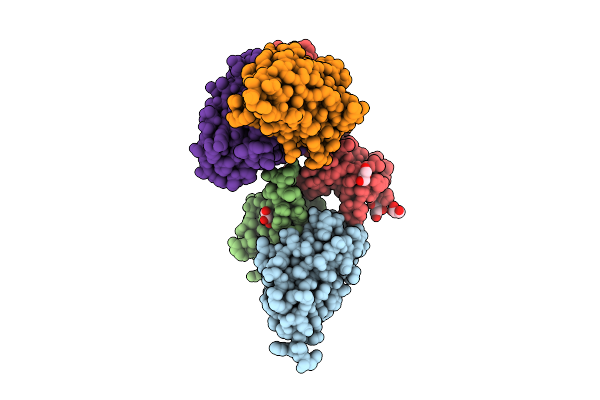 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: CYTOKINE Ligands: GOL |