Search Count: 50,520
All
Selected
 |
Cryo-Em Structure Of D1R In Complex With De Novo Designed Negative Allosteric Gem Targeting Tm5/6/7
Organism: Homo sapiens, Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:2.77 Å Release Date: 2026-03-04 Classification: MEMBRANE PROTEIN Ligands: CLR, A1EKL, PLM |
 |
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-03-04 Classification: ELECTRON TRANSPORT Ligands: CDL, HDD, HEB, MQ9 |
 |
Structural Insights Into Photosystem Ii Supercomplex Of A A Siphonous Green Algae Bryopsis Corticulans From Intertidal Zone
|
 |
Cryo-Em Structure Of Apo Gpr50 With Bril Fusion, Anti-Bril Fab, And Anti-Fab Nb Complex
Organism: Synthetic construct, Homo sapiens, Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:2.98 Å Release Date: 2026-02-18 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-02-18 Classification: RIBOSOME Ligands: ZN, SPD, MG, PUT, K, SPM, NAD, FES, GDP, ATP, VAL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-02-18 Classification: RIBOSOME Ligands: ZN, K, SPD, PUT, MG, VAL, FES, NAD, SPM, ATP, GDP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-02-18 Classification: RIBOSOME Ligands: ZN, K, SPD, PUT, MG, VAL, FES, NAD, SPM, ATP, GDP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.02 Å Release Date: 2026-02-18 Classification: RIBOSOME Ligands: ZN, K, SPD, PUT, MG, FES, NAD, SPM, ATP, GDP, VAL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-02-18 Classification: RIBOSOME Ligands: ZN, K, SPD, PUT, MG, FES, NAD, SPM, ATP, GDP, VAL |
 |
In Situ Structure Of The Human Mitochondrial Large Subunit 39S In Complex With Taco1
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-02-18 Classification: RIBOSOME Ligands: ZN, K, MG, FES, SPD, PUT, VAL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-02-18 Classification: RIBOSOME Ligands: ZN, K, SPD, PUT, MG, FES, NAD, SPM, ATP, GDP, VAL |
 |
Cryo-Em Structure Of The Yeast Respiratory Complex Ii With The Inhibitor Bixafen
Organism: Saccharomyces cerevisiae, Saccharomyces cerevisiae w303
Method: ELECTRON MICROSCOPY Release Date: 2026-02-11 Classification: OXIDOREDUCTASE Ligands: FAD, K, FES, SF4, F3S, A1I6X, 3PE |
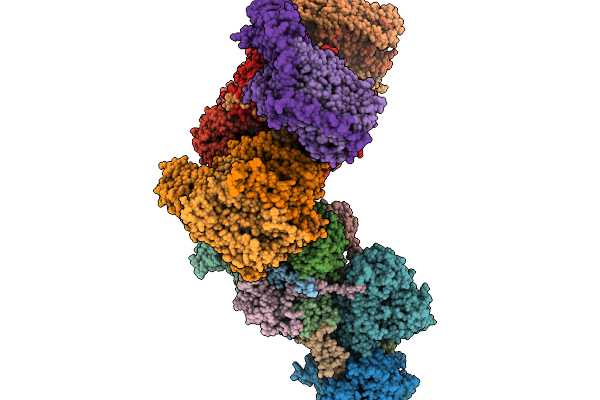 |
Organism: Paracoccus denitrificans pd1222
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: ELECTRON TRANSPORT Ligands: SF4, NA, FES, FMN, DU0, U10, ZN, CDL, HEM, CA, HEC, HEA, CU, MN, CUA |
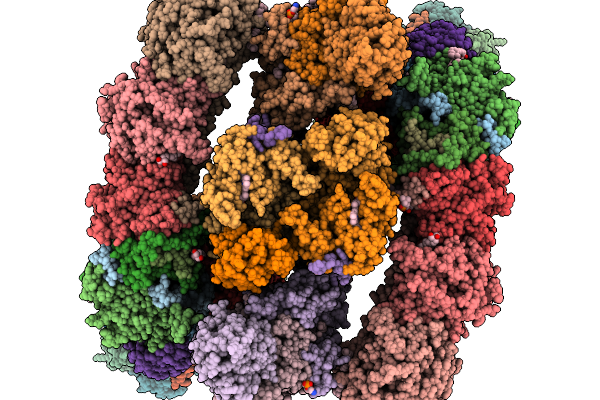 |
Respiratory Supercomplex Ci2-Ciii2-Civ2 (Megacomplex) From Alphaproteobacterium
|
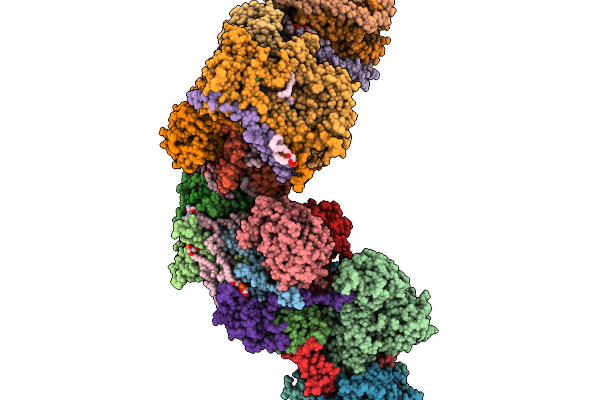 |
Respiratory Supercomplex Ci1-Ciii2-Civ1 (Respirasome) From Alphaproteobacterium
|
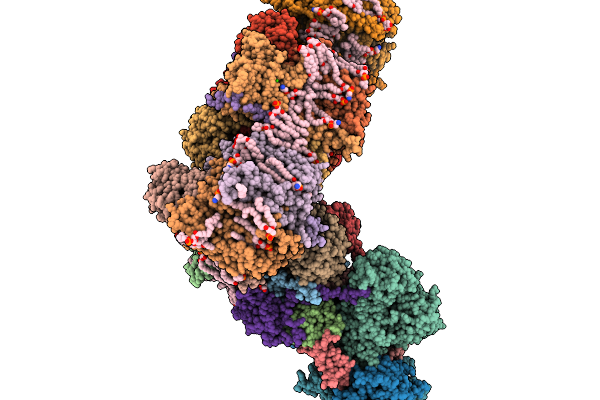 |
 |
Organism: Escherichia coli, Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: SIGNALING PROTEIN Ligands: A1L7J |
 |
Organism: Homo sapiens, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
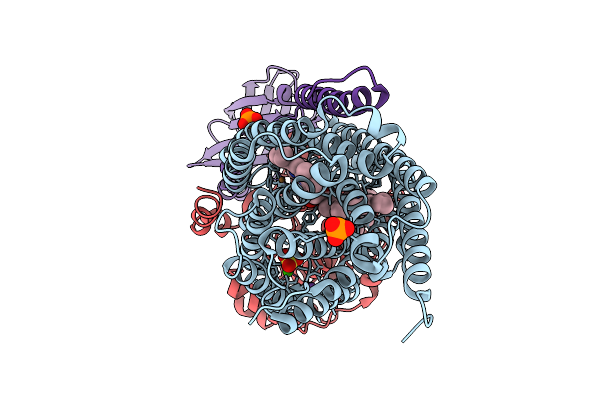 |
Organism: Stutzerimonas stutzeri atcc 14405 = ccug 16156
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2026-01-28 Classification: OXIDOREDUCTASE Ligands: HEM, CU, CA, PO4, NA, SO4, HEC |
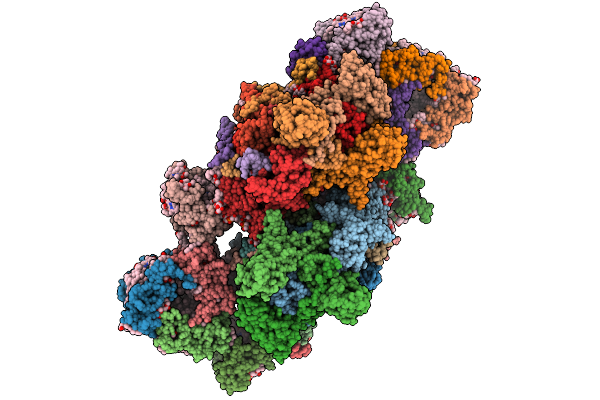 |
Structure Of Unstacked C2S2-Type Psii-Lhcii Supercomplex From Pisum Sativum
|

