Search Count: 186
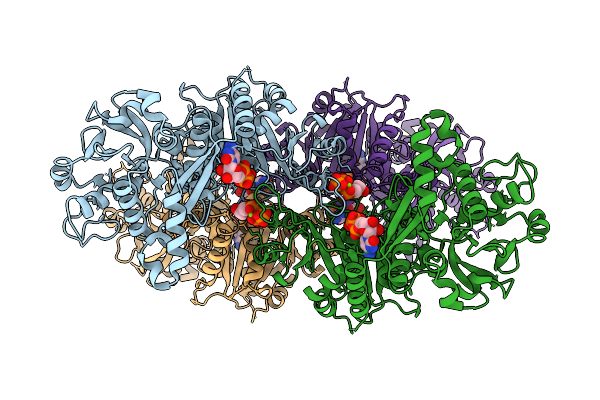 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: STRUCTURAL PROTEIN Ligands: CTP, MG |
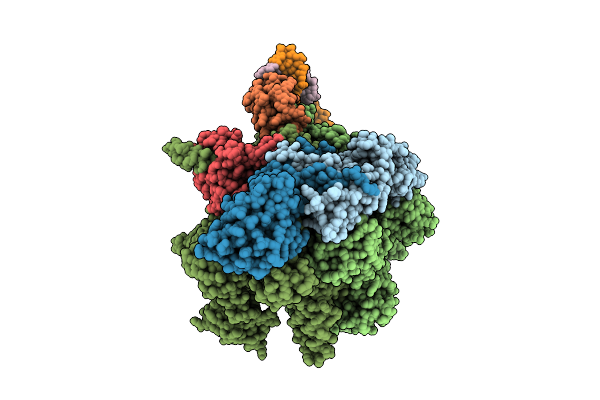 |
Cryoem Structure Of The T.Thermophilus Transcription Initiation Complex Bound To Gp4A-C-U And Ctp, -1 Dc In The Template Dna Strand
Method: ELECTRON MICROSCOPY
Release Date: 2025-10-22 Classification: TRANSCRIPTION Ligands: MG, 1N7, CTP, ZN |
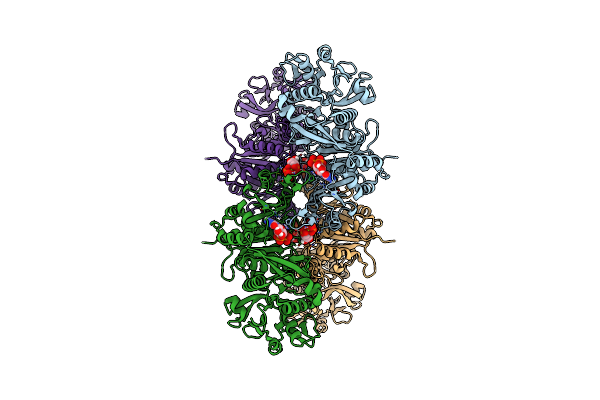 |
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: LIGASE Ligands: CTP, MG |
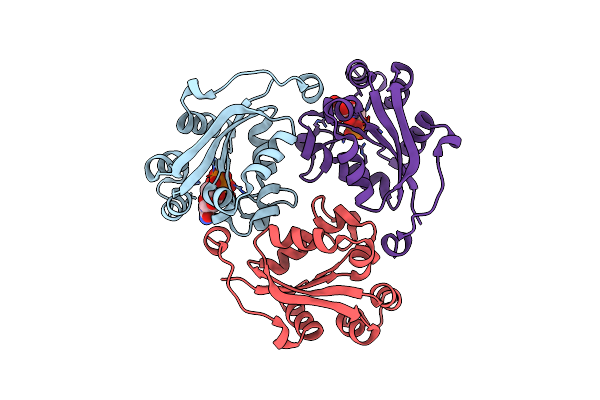 |
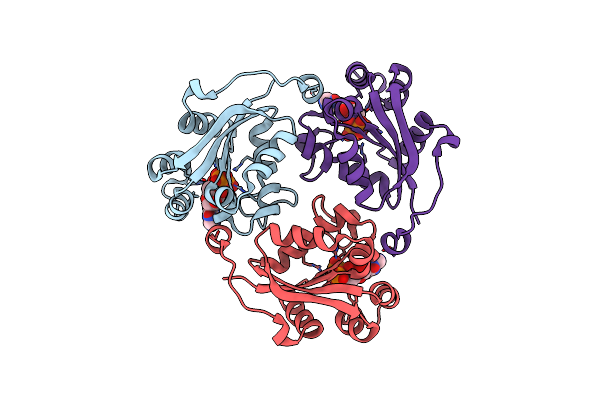
Crystal Structure Of Nucleoside-Diphosphate Kinase From Cryptosporidium Parvum In Complex With Cytidine-5'-Diphosphate And Cytidine-5'-Triphosphate
Organism: Cryptosporidium parvum iowa ii
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2025-04-30 Classification: TRANSFERASE Ligands: CTP, MG, CDP |
 |
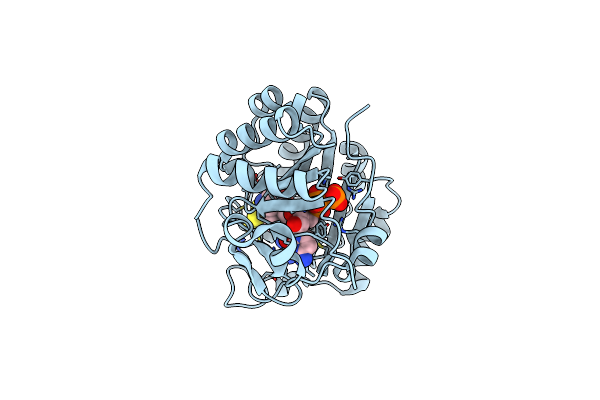
Crystal Structure Of Nucleoside-Diphosphate Kinase From Cryptosporidium Parvum In Complex With With Adp And Ctp
Organism: Cryptosporidium parvum iowa ii
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-04-30 Classification: TRANSFERASE Ligands: CTP, ADP, MG |
 |
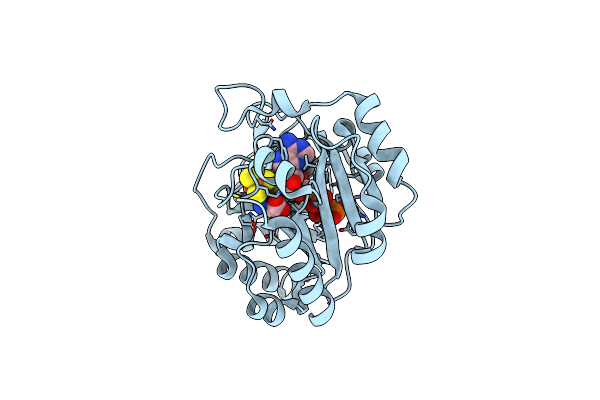
X-Ray Crystal Structure Of The Second Viperin-Like Enzyme From Trichoderma Virens With Bound Ctp And Sam
Organism: Trichoderma virens
Method: X-RAY DIFFRACTION Release Date: 2025-02-19 Classification: ANTIVIRAL PROTEIN Ligands: SF4, SAM, CTP, CL |
 |
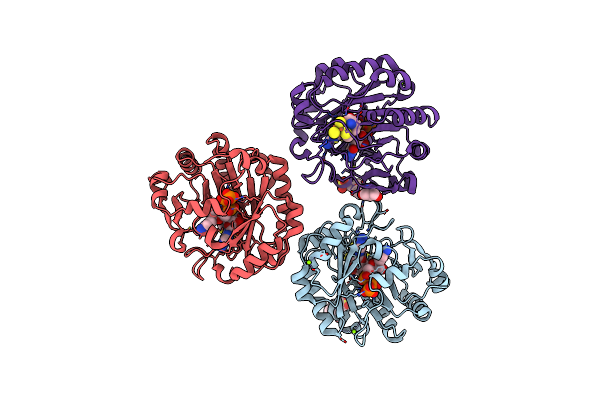
X-Ray Crystal Structure Of The Second Viperin-Like Enzyme From T. Virens Variant F40H With Bound Ctp And Sam
Organism: Trichoderma virens
Method: X-RAY DIFFRACTION Release Date: 2025-02-19 Classification: ANTIVIRAL PROTEIN Ligands: CTP, SF4, SAM, CL |
 |
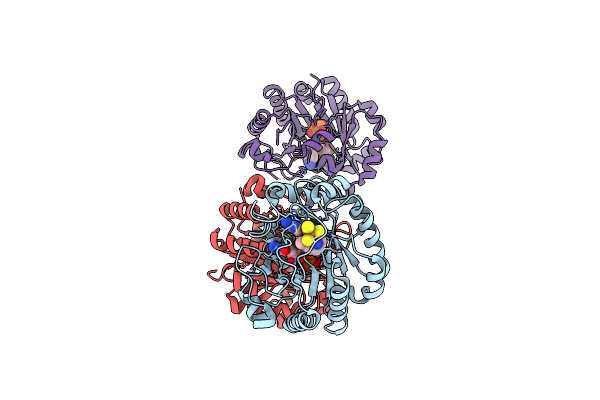
X-Ray Crystal Structure Of An Engineered Viperin-Like Enzyme From T. Virens With Bound Ctp And Sam
Organism: Trichoderma virens
Method: X-RAY DIFFRACTION Release Date: 2025-02-12 Classification: ANTIVIRAL PROTEIN Ligands: SF4, SAM, CTP, EPE, MG, GOL |
 |
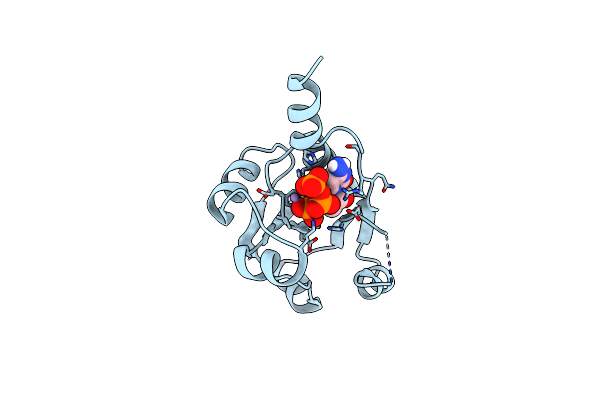
X-Ray Crystal Structure Of The Viperin-Like Enzyme From T. Virens With Bound Ctp And Sam
Organism: Trichoderma virens
Method: X-RAY DIFFRACTION Release Date: 2025-02-12 Classification: ANTIVIRAL PROTEIN Ligands: CTP, SF4, SAM |
 |
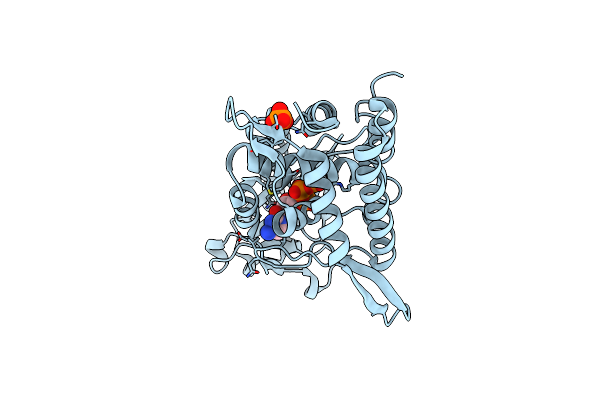
Crystal Structure Of C-Terminal Catalytic Domain Of Plasmodium Falciparum Ctp:Phosphocholine Cytidylyltransferase With Cytidine-Triphosphate
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-02-05 Classification: TRANSFERASE Ligands: CTP, MN |
 |
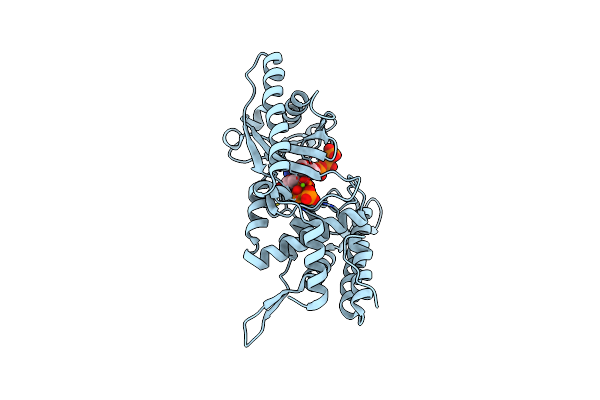
Crystal Structure Of The Deinococcus Wulumuqiensis Cd-Ntase Dwcdnb In Complex With Ctp Mn
Organism: Deinococcus wulumuqiensis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-01-29 Classification: TRANSFERASE Ligands: CTP, MN, PO4 |
 |
Crystal Structure Of The Deinococcus Wulumuqiensis Cd-Ntase Dwcdnb In Complex With Ctp
Organism: Deinococcus wulumuqiensis
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2025-01-29 Classification: TRANSFERASE Ligands: CTP, MG |
 |
Replication-Like Initiation State Of Influenza Polymerase With Gtp And Ctp At Respectively The -1 And +1 Positions (Strain A/Little Yellow-Shouldered Bat/Guatemala/060/2010/H17N10)
Organism: Influenza a virus
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2024-09-18 Classification: VIRAL PROTEIN Ligands: GOL, CTP, GTP, MG, MGT |
 |
Organism: Monkeypox virus, Dna molecule
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: VIRAL PROTEIN/DNA Ligands: MG, CTP |
 |
Human Dna Polymerase Eta-Dna-Dt Primer Rctp Insertion Ternary Complex At Ph7.0 (K+ Mes) With 1 Ca2+ Ion
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2024-05-15 Classification: Transferase/DNA Ligands: GOL, CTP, CA, K |
 |
Crystal Structure Of Mycobacterium Smegmatis Coab In Complex With Ctp And 5-Methoxy-1H-Indole-2-Carboxylic Acid
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:2.84 Å Release Date: 2024-05-08 Classification: LIGASE Ligands: CTP, CA, VEO |
 |
Crystal Structure Of Mycobacterium Smegmatis Coab In Complex With Ctp And 2-(5-Bromo-1-(4-Hydroxyphenyl)-1H-Indol-3-Yl)-2-Oxoacetic Acid
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2024-05-08 Classification: LIGASE Ligands: CTP, CA, VE6 |
 |
Crystal Structure Of Mycobacterium Smegmatis Coab In Complex With Ctp And 2-(1-(4-Hydroxyphenyl)-5-Phenyl-1H-Indol-3-Yl)-2-Oxoacetic Acid
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2024-05-08 Classification: LIGASE Ligands: CTP, CA, W52, ACT |
 |
Crystal Structure Of Mycobacterium Smegmatis Coab In Complex With Ctp And 2-(1-(3,4-Dihydroxyphenyl)-5-Phenyl-1H-Indol-3-Yl)-2-Oxoacetic Acid
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2024-05-08 Classification: LIGASE Ligands: CTP, CA, W4X |
 |
Crystal Structure Of Mycobacterium Smegmatis Coab In Complex With Ctp And 2-(1-(3,4-Dihydroxyphenyl)-5-(3-Hydroxyphenyl)-1H-Indol-3-Yl)-2-Oxoacetic Acid
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2024-05-08 Classification: LIGASE Ligands: CTP, CA, W5E, ACT |

