Search Count: 10
All
Selected
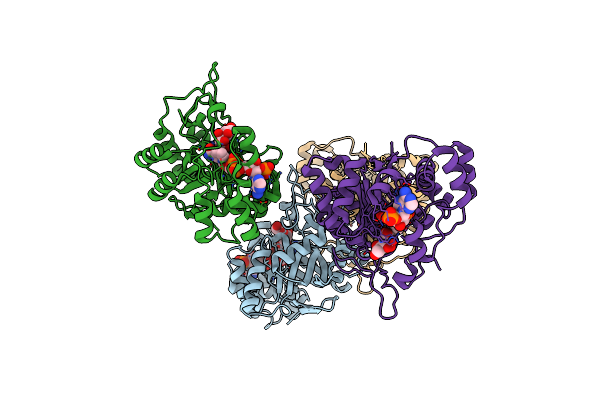 |
Crystal Structure Of Mannose-6-Phosphate Reductase From Celery (Apium Graveolens) Leaves With Nadp+ And Mannonic Acid Bound
Organism: Apium graveolens
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2022-03-16 Classification: OXIDOREDUCTASE Ligands: NAP, CS2 |
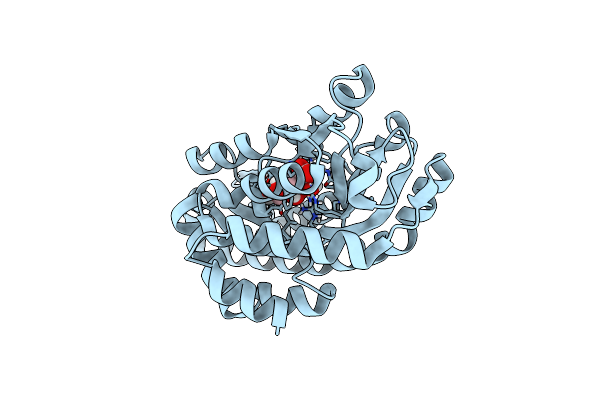 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Rosenbacter Denitrificans Och 114 (Rd1_1052, Target Efi-510238) With Bound D-Mannonate
Organism: Roseobacter denitrificans
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2014-05-21 Classification: SOLUTE-BINDING PROTEIN Ligands: CS2 |
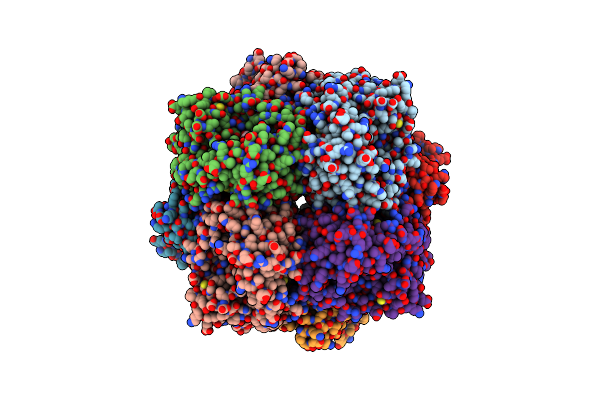 |
Crystal Structure Of D-Mannonate Dehydratase From Chromohalobacter Salexigens Complexed With Mg,D-Mannonate And 2-Keto-3-Deoxy-D-Gluconate
Organism: Chromohalobacter salexigens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-05-14 Classification: HYDROLASE Ligands: CS2, CL, MG, KDG, GOL |
 |
Crystal Structure Of The A314P Mutant Of Mannonate Dehydratase From Novosphingobium Aromaticivorans Complexed With Mg And D-Mannonate
Organism: Novosphingobium aromaticivorans
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2014-04-09 Classification: ISOMERASE Ligands: MG, CS2, PGE, CO2, PDO, CO3 |
 |
Crystal Structures Of Mannonate Dehydratase From Escherichia Coli Strain K12 Complexed With D-Mannonate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2013-03-27 Classification: LYASE Ligands: CL, CS2, MN |
 |
Crystal Structure Of Fructuronate Reductase (Ydfi) From E. Coli Cft073 (Efi Target Efi-506389) Complexed With Nadh And D-Mannonate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-01-23 Classification: OXIDOREDUCTASE Ligands: CS2, NAI, SO4 |
 |
Crystal Structure Of Mannonate Dehydratase (Target Efi-502209) From Caulobacter Crescentus Cb15 Complexed With Magnesium And D-Mannonate
Organism: Caulobacter crescentus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-09-12 Classification: LYASE Ligands: MG, CL, GOL, CO3, CS2 |
 |
Crystal Structure Of D-Mannonate Dehydratase From Chromohalobacter Salexigens Complexed With Mg,D-Mannonate And 2-Keto-3-Deoxy-D-Gluconate
Organism: Chromohalobacter salexigens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-10-19 Classification: ISOMERASE Ligands: MG, CS2, KDG |
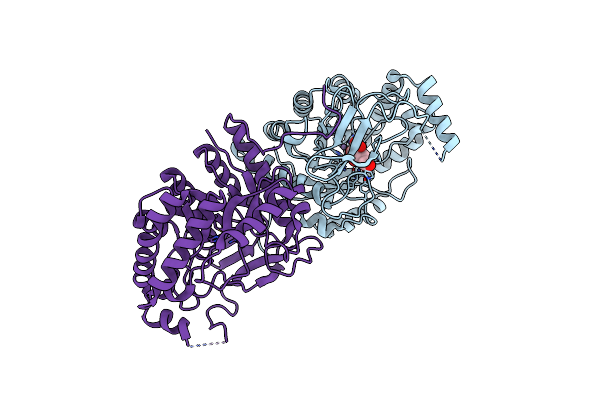 |
Crystal Structure Of The Streptoccocus Suis Serotype2 D-Mannonate Dehydratase In Complex With Its Substrate
Organism: Streptococcus suis
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2009-06-23 Classification: LYASE Ligands: MN, CS2 |
 |
Crystal Structure Of The K271E Mutant Of Mannonate Dehydratase From Novosphingobium Aromaticivorans Complexed With Mg And D-Mannonate
Organism: Novosphingobium aromaticivorans
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2007-10-30 Classification: LYASE Ligands: MG, CS2 |

