Search Count: 400
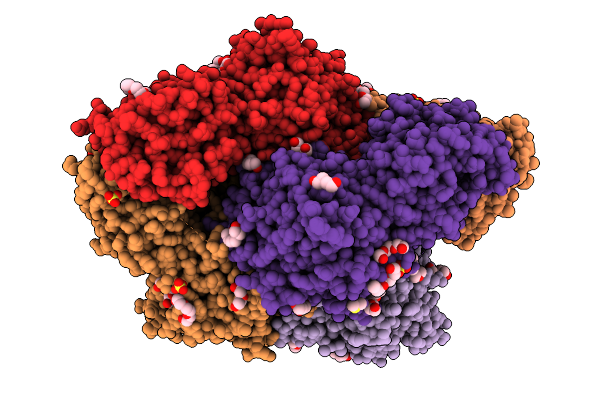 |
Plasmodium Falciparum M17 Aminopeptidase (Pfa-M17) Bound To Inhibitor 3Ab (Mips3413)
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2026-01-28 Classification: HYDROLASE Ligands: ZN, CO3, PEG, SO4, A1CT4, 1PE, DMS, 2PE, A1CT5 |
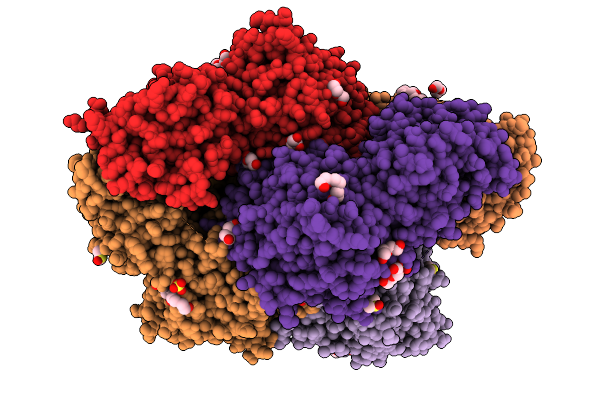 |
Plasmodium Falciparum M17 Aminopeptidase (Pfa-M17) Bound To Inhibitor 3K (Mips3415)
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2026-01-28 Classification: HYDROLASE Ligands: ZN, CO3, PEG, DMS, SO4, GOL, A1CTH, 1PE |
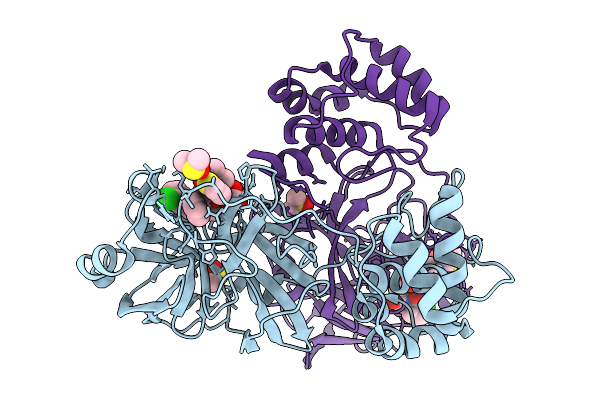 |
Crystal Structure Of Sars-Cov-2 Main Protease (Mpro) In Complex With The Covalently Bound Inhibitor Gue-4303 (Compound 12 In Publication)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2026-01-14 Classification: ANTIVIRAL PROTEIN Ligands: A1JNF, DMS, CO3 |
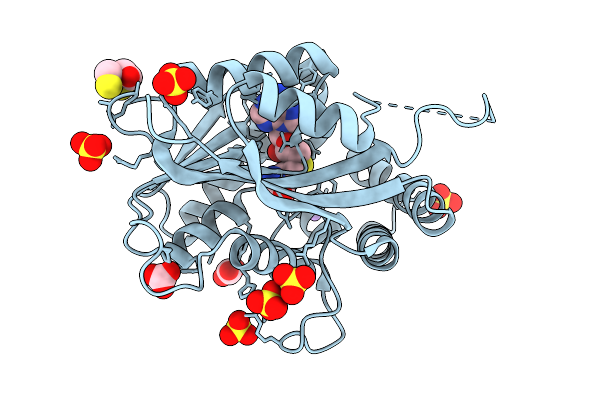 |
Organism: Burkholderia territorii
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2025-12-31 Classification: BIOSYNTHETIC PROTEIN Ligands: SAH, DTT, CO3, SO4, NA |
 |
T-Muurolol Synthase From Roseiflexus Castenholzii (Tms) In Complex With Fspp
Organism: Roseiflexus castenholzii
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2025-12-24 Classification: LYASE Ligands: MG, FPS, GOL, CO3, SO4, EDO |
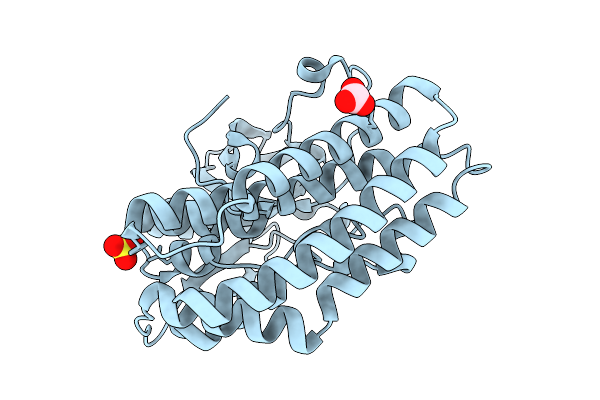 |
Crystal Structure Of Ctag From Ruminiclostridium Cellulolyticum (P2(1)-Small)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2025-10-01 Classification: LIGASE Ligands: SO4, CO3 |
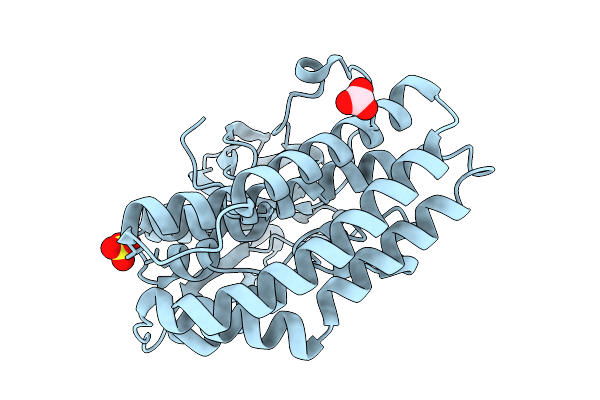 |
Crystal Structure Of The Ctag_H128A Variant From Ruminiclostridium Cellulolyticum (P2(1)-Small)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-10-01 Classification: LIGASE Ligands: SO4, CO3 |
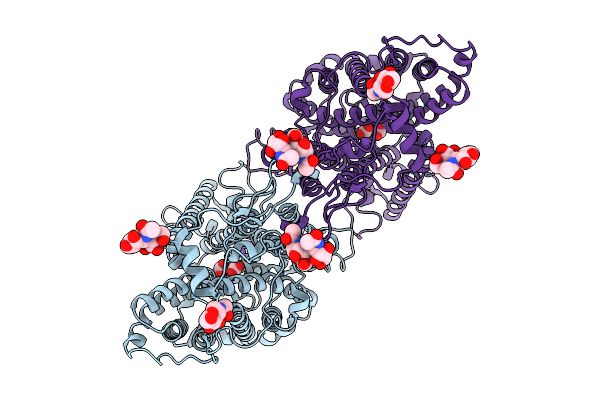 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.30 Å Release Date: 2025-09-03 Classification: PROTEIN TRANSPORT Ligands: CO3, NAG, NA |
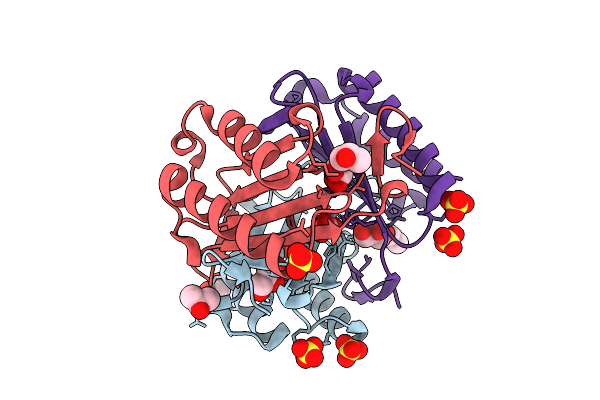 |
Macrophage Migration Inhibitory Factor S61H/Y100H Mutant Complexed With Three Zinc Ions (Zn3-Mif(S61H/Y100H)-L)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2025-08-06 Classification: CYTOKINE Ligands: ZN, CO3, SO4, GOL, IPA |
 |
Macrophage Migration Inhibitory Factor S61H/Y100H Mutant Complexed With Three Zinc Ions (Zn3-Mif(S61H/Y100H))
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2025-08-06 Classification: CYTOKINE Ligands: ZN, CO3, SO4, GOL, IPA |
 |
Macrophage Migration Inhibitory Factor Y100H Mutant Complexed With Three Zinc Ions (Zn3-Mif(Y100H))
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2025-08-06 Classification: CYTOKINE Ligands: ZN, CL, CO3, SO4, GOL, IPA |
 |
Crystal Structure Of The Immunity Protein (52 Domain-Containing Protein) From Pseudomonas Aeruginosa Pao1.
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2025-06-18 Classification: ANTITOXIN Ligands: EPE, PO4, EDO, CO3 |
 |
Structure Of Human Butyrylcholinesterase With 3-(((2-Cycloheptylethyl)(Methyl)Amino)Methyl)-N-Methyl-1H-Indol-7-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2024-10-23 Classification: HYDROLASE Ligands: GOL, NAG, MES, WXN, CO3, SO4 |
 |
Organism: Kitasatospora cystarginea
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2024-08-21 Classification: TRANSFERASE Ligands: SAH, CA, SO4, CO3, EDO, PG4 |
 |
Crystal Structure Of Cis-Epoxysuccinate Hydrolase From Klebsiella Oxytoca With L(+)-Tartaric Acid
Organism: Klebsiella oxytoca
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2024-06-12 Classification: HYDROLASE Ligands: TLA, EDO, GOL, CO3 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-22 Classification: TRANSLOCASE Ligands: NAG, PIO, PLC, CO3 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-22 Classification: TRANSLOCASE Ligands: PIO, PLC, CO3, NAG |
 |
Organism: Salmonella enterica subsp. enterica serovar typhi str. ct18
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2024-05-08 Classification: ISOMERASE Ligands: CO3 |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2024-05-01 Classification: CHAPERONE Ligands: 1PE, CO3, GOL, SO4 |
 |
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-04-24 Classification: HYDROLASE Ligands: CO3, ZN, WRC, 1PE, SO4 |

