Search Count: 19,908
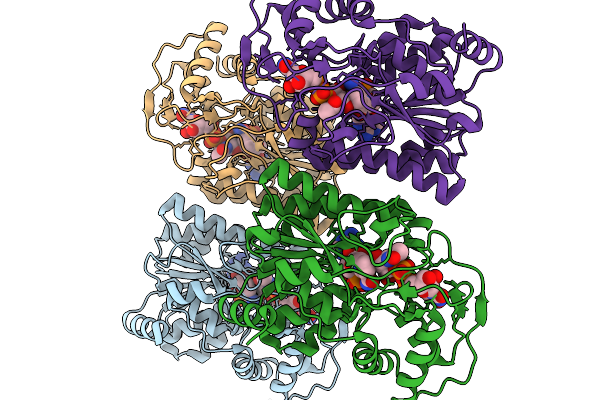 |
Crystal Structure Of Capsular Polysaccharide Biosynthesis Protein From Bordetella Pertussis In Complex With Nad And Uridine-Diphosphate-N-Acetylgalactosamine (Cocrystallization)
Organism: Bordetella pertussis tohama i
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2026-02-04 Classification: ISOMERASE Ligands: CL, NAD, UD2 |
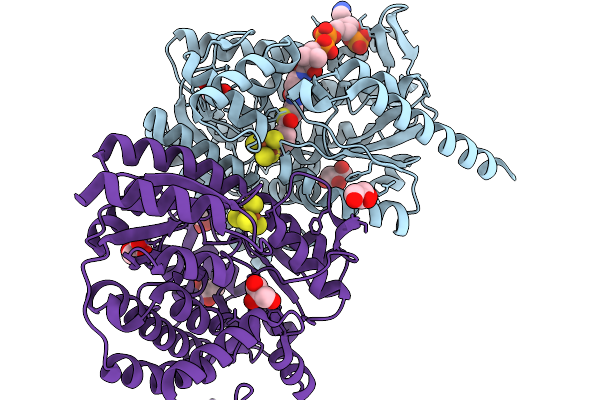 |
3-Methylbenzoyl-Coa Reductase From Thauera Chlorobenzoica (Subunits Mbdon )
Organism: Thauera chlorobenzoica
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2026-02-04 Classification: OXIDOREDUCTASE Ligands: SF4, BYC, GOL, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2026-02-04 Classification: ANTIVIRAL PROTEIN Ligands: CL, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.83 Å Release Date: 2026-02-04 Classification: ANTIVIRAL PROTEIN Ligands: SO4, GOL, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2026-02-04 Classification: ANTIVIRAL PROTEIN Ligands: CL, SO4, ACT |
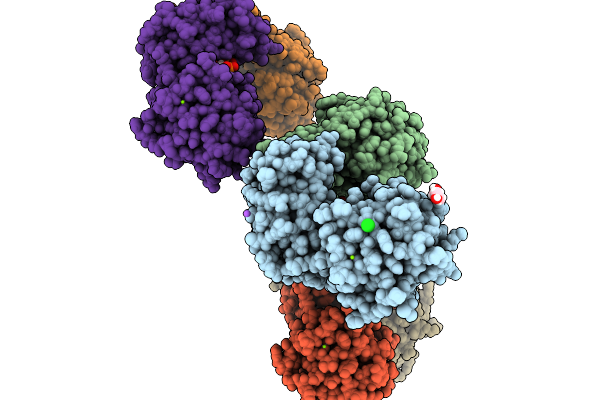 |
Mouse Phosphomannomutase 2 In Complex With The Activator Glucose-1,6-Bisphosphate
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2026-02-04 Classification: ISOMERASE Ligands: GOL, MG, CL, NA, G16 |
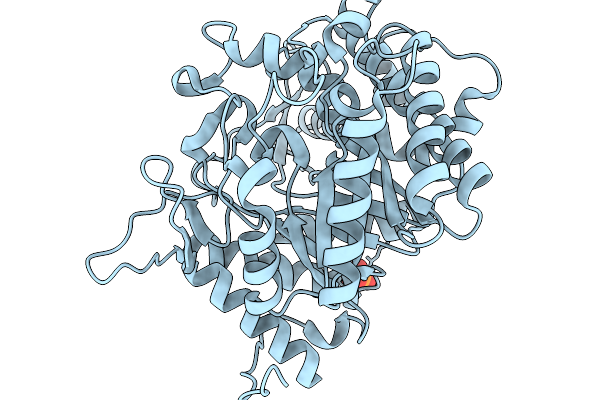 |
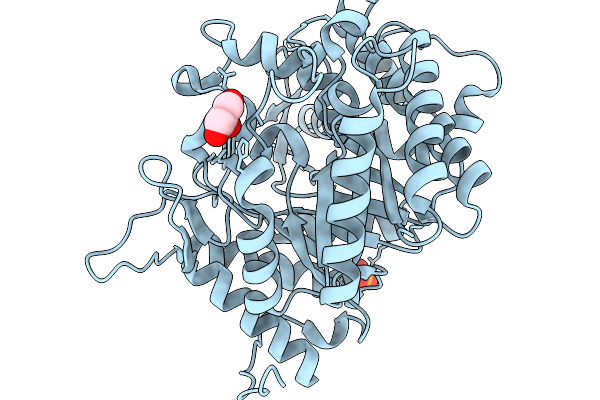
Crystal Structure Of Glucose Tolerant Beta-Glucosidase Unbgl1 Mutant (C188V)
Organism: Soil metagenome
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: CL, PO4 |
 |
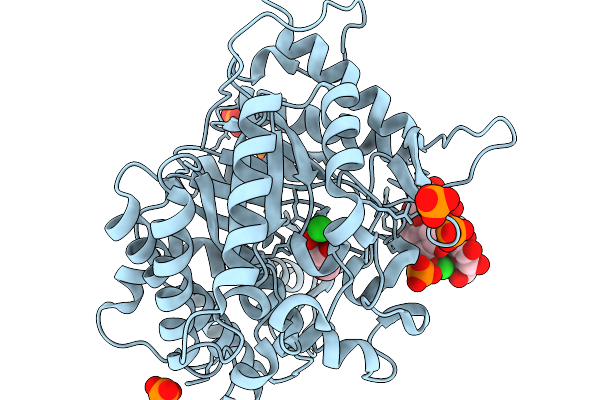
Crystal Structure Of Glucose Tolerant Gh1 Beta-Glucosidase Mutant (Unbgl1_H261W)
Organism: Soil metagenome
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: PO4, GOL, CL |
 |
Crystal Structure Of Cellobiose And Glucose Bound Glucose Toleranant Beta-Glucosidase Mutant (Unbgl1_H261W)
Organism: Soil metagenome
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: BGC, PO4, CL |
 |
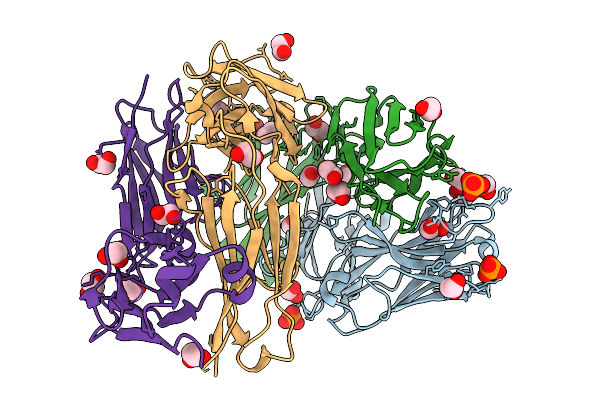
High Resolution Crystal Structure Of Gh1 Beta-Glucosidase From Soil Metagenome (Unbgl1)
Organism: Soil metagenome
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: PO4, CL, GOL |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2026-02-04 Classification: IMMUNE SYSTEM Ligands: EDO, CL, PO4 |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2026-02-04 Classification: UNKNOWN FUNCTION Ligands: CL |
 |
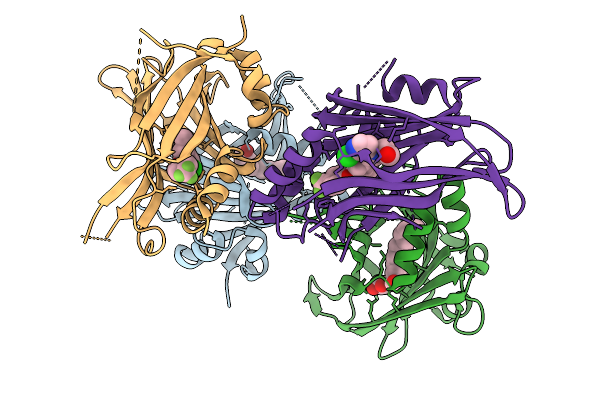
Organism: Homo sapiens, Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2026-02-04 Classification: IMMUNE SYSTEM Ligands: EDO, PGE, CL, PEG, TRS, ACT, CA, CAC |
 |
Human Tead1 In Complex With 2-(4-Chloro-3-{3-Methyl-5-[4-(Trifluoromethyl)Phenoxy]Phenyl}-1H-Pyrrolo[3,2-C]Pyridin-1-Yl)Ethan-1-Ol
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2026-02-04 Classification: TRANSCRIPTION Ligands: MYR, GOL, CL, A1JLZ |
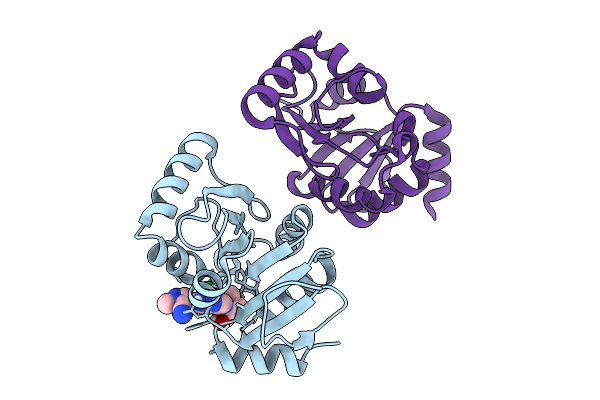 |
Pandda Analysis Group Deposition -- Crystal Structure Of Sars-Cov-2 Nsp3 Macrodomain In Complex With Avi-0004054
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.02 Å Release Date: 2026-01-28 Classification: VIRAL PROTEIN Ligands: A1CXU, CL |
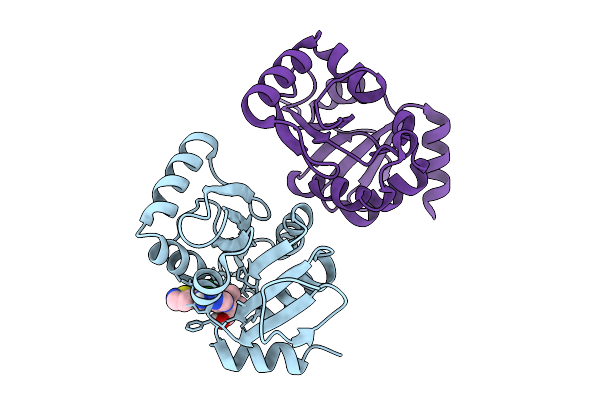 |
Pandda Analysis Group Deposition -- Crystal Structure Of Sars-Cov-2 Nsp3 Macrodomain In Complex With Avi-0004100
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.01 Å Release Date: 2026-01-28 Classification: VIRAL PROTEIN Ligands: A1CXV, CL |
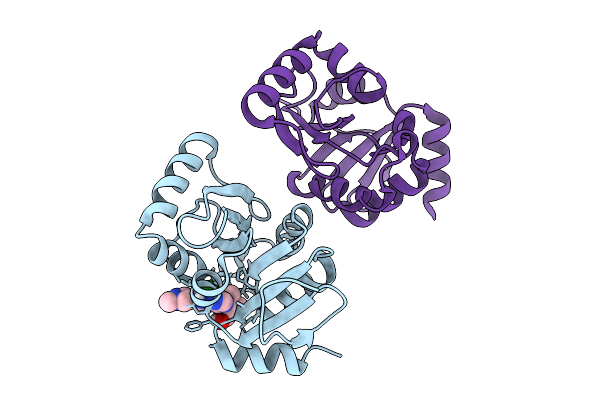 |
Pandda Analysis Group Deposition -- Crystal Structure Of Sars-Cov-2 Nsp3 Macrodomain In Complex With Avi-0004214
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.01 Å Release Date: 2026-01-28 Classification: VIRAL PROTEIN Ligands: A1CXW, CL |
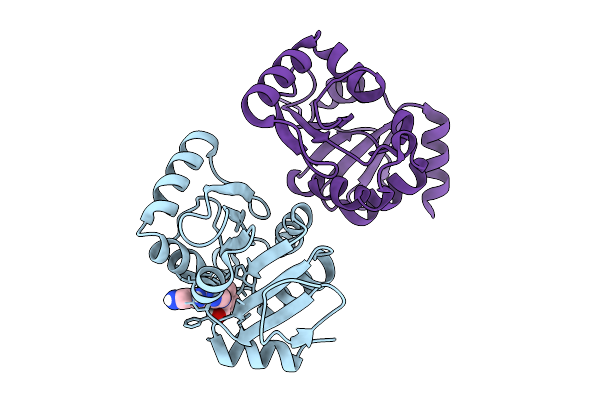 |
Pandda Analysis Group Deposition -- Crystal Structure Of Sars-Cov-2 Nsp3 Macrodomain In Complex With Avi-0004268
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:0.95 Å Release Date: 2026-01-28 Classification: VIRAL PROTEIN Ligands: A1CXX, CL |
 |
Pandda Analysis Group Deposition -- Crystal Structure Of Sars-Cov-2 Nsp3 Macrodomain In Complex With Avi-0004683
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:0.99 Å Release Date: 2026-01-28 Classification: VIRAL PROTEIN Ligands: A1CXZ, CL |
 |
Pandda Analysis Group Deposition -- Crystal Structure Of Sars-Cov-2 Nsp3 Macrodomain In Complex With Avi-0004678
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:0.99 Å Release Date: 2026-01-28 Classification: VIRAL PROTEIN Ligands: A1CX0, CL |

