Search Count: 164
All
Selected
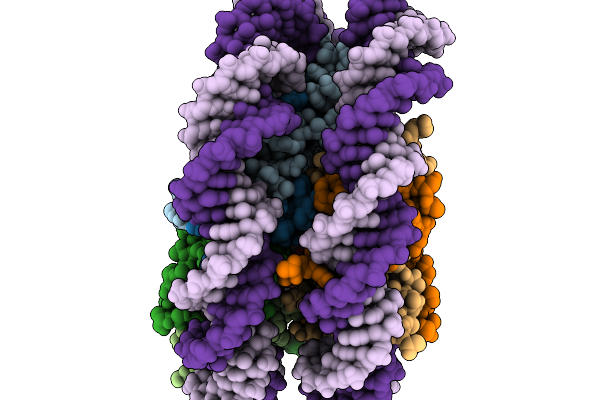 |
Organism: Schizosaccharomyces pombe (strain 972 / atcc 24843)
Method: ELECTRON MICROSCOPY Resolution:2.99 Å Release Date: 2026-02-04 Classification: NUCLEAR PROTEIN/DNA |
 |
Organism: Schizosaccharomyces pombe (strain 972 / atcc 24843)
Method: ELECTRON MICROSCOPY Resolution:3.04 Å Release Date: 2026-02-04 Classification: NUCLEAR PROTEIN/DNA |
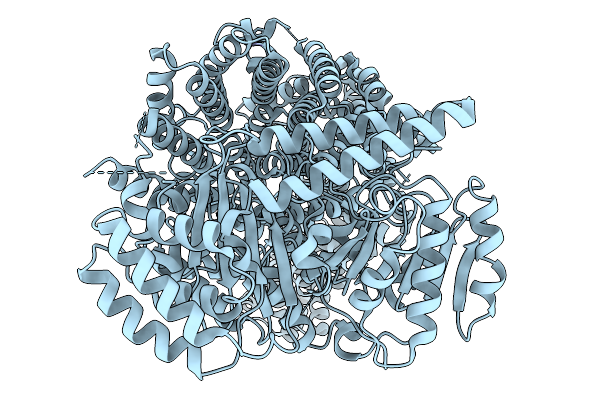 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.50 Å Release Date: 2026-01-14 Classification: CELL CYCLE Ligands: ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.30 Å Release Date: 2025-09-03 Classification: CELL CYCLE Ligands: ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.40 Å Release Date: 2025-09-03 Classification: CELL CYCLE Ligands: ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: CELL CYCLE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.80 Å Release Date: 2025-09-03 Classification: CELL CYCLE Ligands: ZN |
 |
Cryo-Em Structure Of Human Separase Bound To Phosphorylated Scc1 (310-550 Aa)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.93 Å Release Date: 2025-09-03 Classification: CELL CYCLE Ligands: ZN |
 |
Cryo-Em Structure Of Human Separase Bound To Phosphorylated Scc1 (100-320 Aa)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.96 Å Release Date: 2025-09-03 Classification: CELL CYCLE Ligands: ZN |
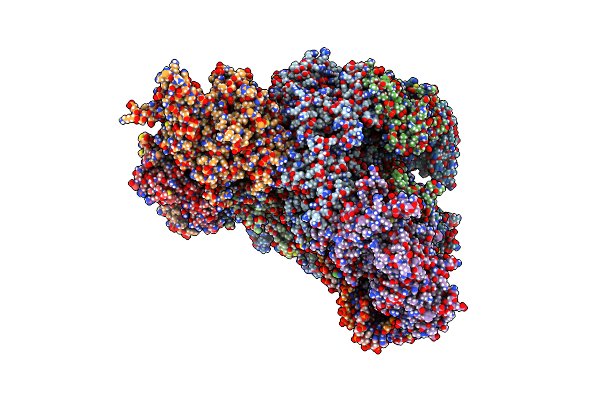 |
Cryo-Em Structure Of Yeast Cmg Helicase Stalled At G4-Containing Dna Template, State 1
Organism: Saccharomyces cerevisiae w303, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: REPLICATION/DNA Ligands: ADP, MG, ZN, ATP |
 |
Organism: Photorhabdus thracensis, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: DNA BINDING PROTEIN Ligands: MG, ATP |
 |
Organism: Photorhabdus thracensis, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: DNA BINDING PROTEIN Ligands: MG, ATP |
 |
Organism: Photorhabdus thracensis, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: DNA BINDING PROTEIN Ligands: MG, ATP |
 |
Organism: Photorhabdus thracensis, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: DNA BINDING PROTEIN Ligands: MG, ATP |
 |
Organism: Photorhabdus thracensis, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: DNA BINDING PROTEIN Ligands: MG, ATP |
 |
Organism: Escherichia coli, Escherichia phage t7
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: DNA BINDING PROTEIN |
 |
Organism: Escherichia coli, Escherichia phage t7
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: DNA BINDING PROTEIN |
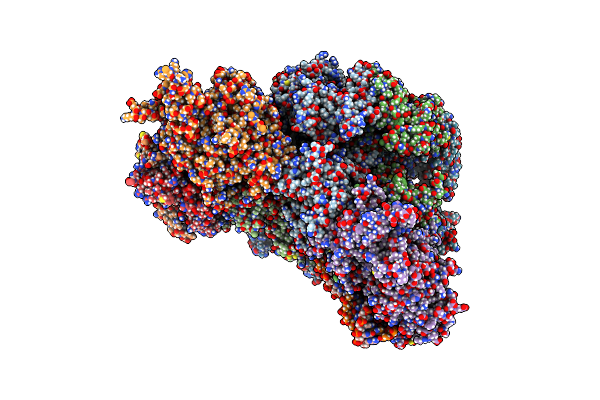 |
Cryo-Em Structure Of Yeast Cmg Helicase Stalled At G4-Containing Dna Template, State 2
Organism: Saccharomyces cerevisiae w303, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: REPLICATION/DNA Ligands: ATP, MG, ZN, ADP |
 |
Chromosome Passenger Complex (Cpc) Localization Module In Complex With H3.T3P-Nucleosome
Organism: Xenopus laevis, Synthetic construct, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: CELL CYCLE Ligands: ZN |
 |
Organism: Xenopus laevis, Unidentified, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: CELL CYCLE |

