Search Count: 22
 |
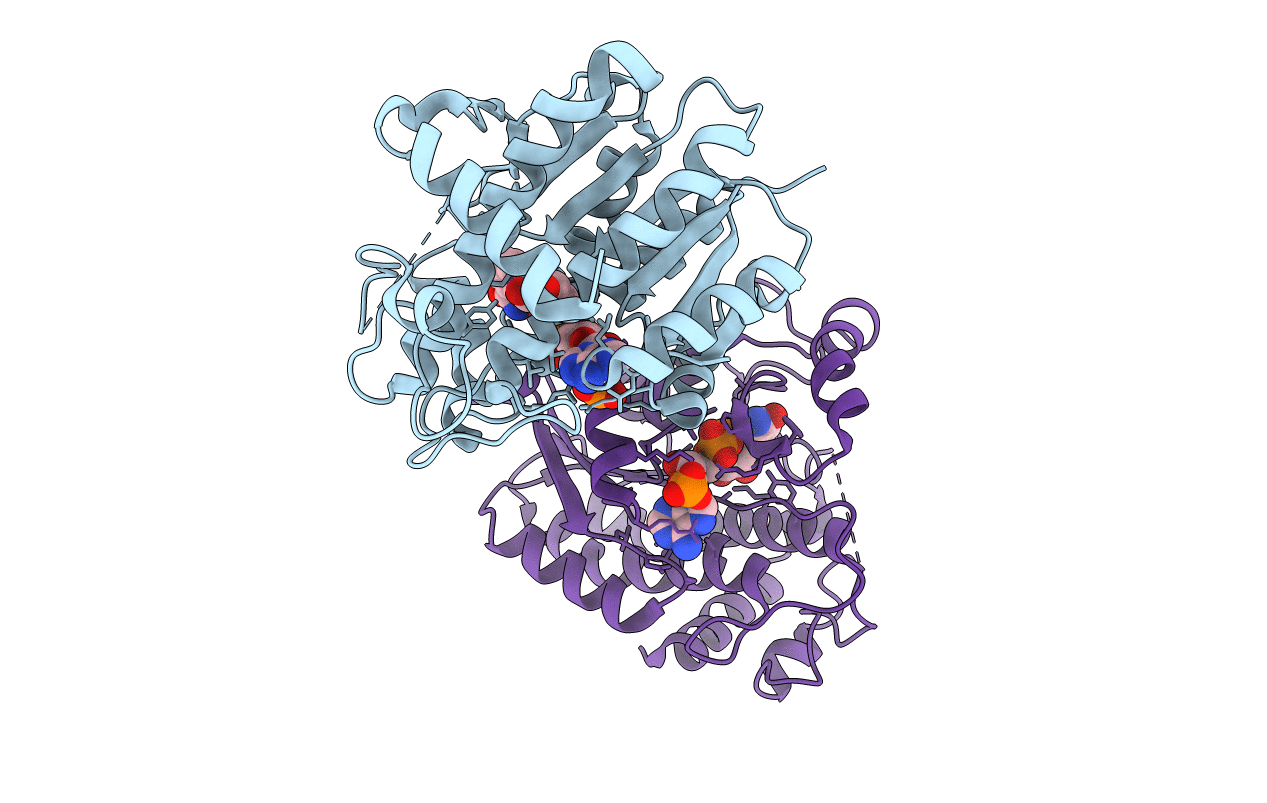
Crystal Structure Of Light-Dependent Protochlorophyllide Oxidoreductase From Synechocystis Sp. Pcc 6803
Organism: Synechocystis sp. (strain pcc 6803 / kazusa)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-04-01 Classification: OXIDOREDUCTASE Ligands: NDP, SO4 |
 |
Crystal Structure Of Light-Dependent Protochlorophyllide Oxidoreductase From Thermosynechococcus Elongatus
Organism: Thermosynechococcus elongatus (strain bp-1)
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2020-04-01 Classification: OXIDOREDUCTASE Ligands: NDP |
 |
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2020-01-15 Classification: HYDROLASE |
 |
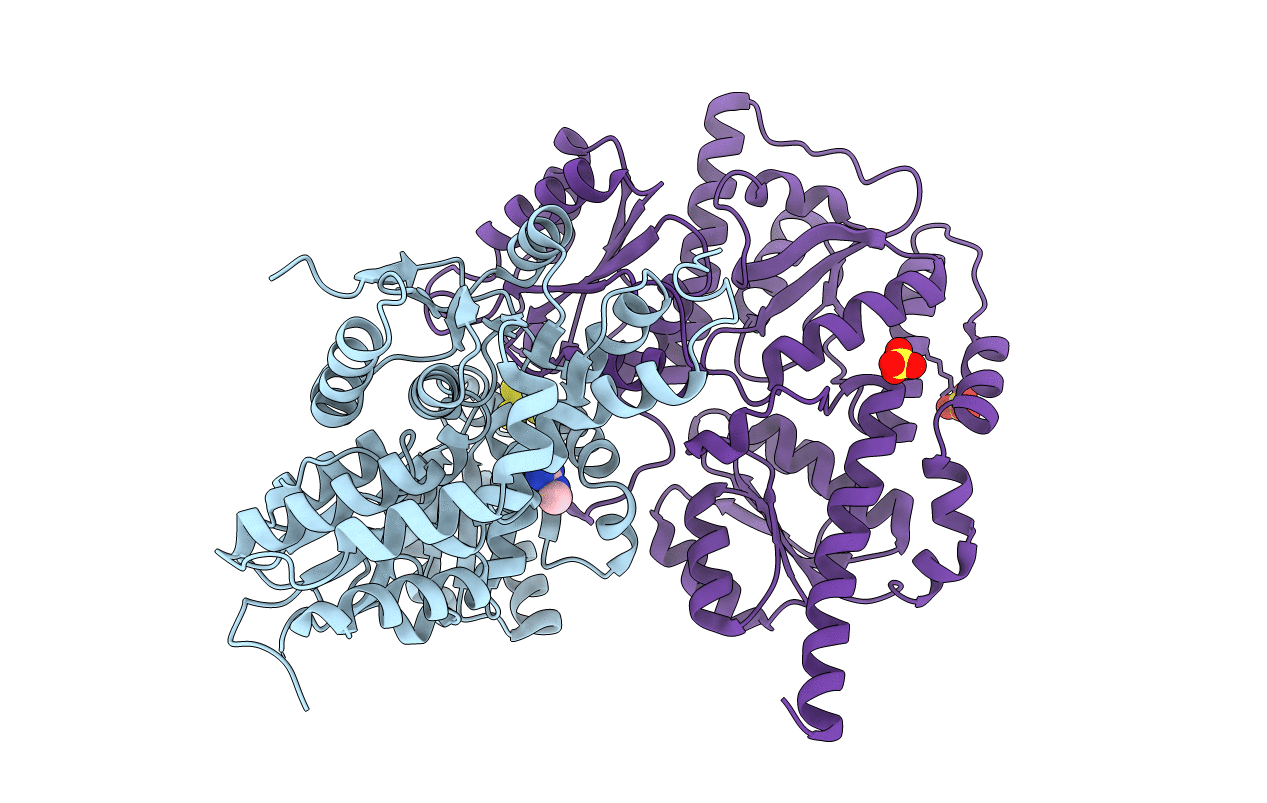
Modelling Of The Complex Between Subunits Bchi And Bchd Of Magnesium Chelatase Based On Single-Particle Cryo-Em Reconstruction At 7.5 Ang
Organism: Rhodobacter capsulatus
Method: ELECTRON MICROSCOPY Resolution:7.50 Å Release Date: 2010-11-10 Classification: LIGASE |
 |
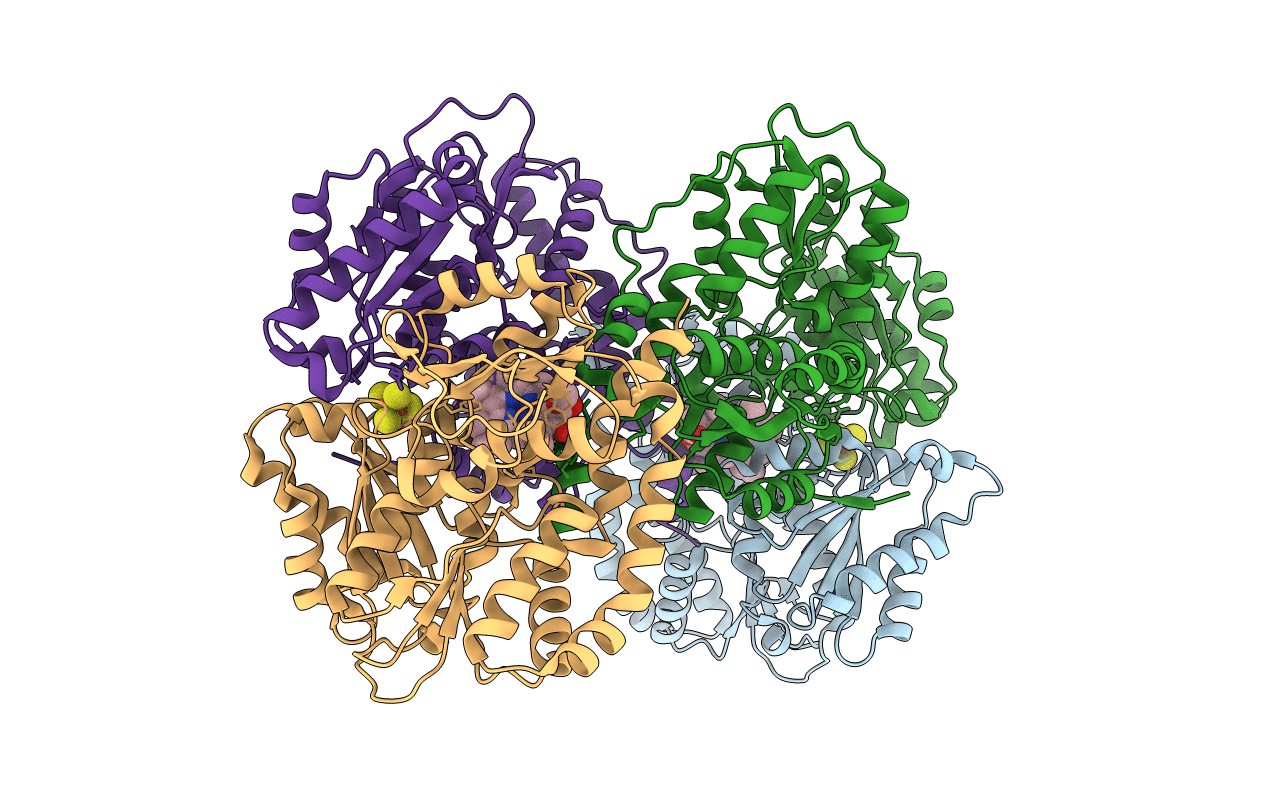
Organism: Thermosynechococcus elongatus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2010-06-16 Classification: OXIDOREDUCTASE Ligands: SF4, MGX, SO4 |
 |
Structure Of The Light-Independent Protochlorophyllide Reductase Catalyzing A Key Reduction For Greening In The Dark
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-04-21 Classification: OXIDOREDUCTASE Ligands: SF4, PMR |
 |
Structure Of The Light-Independent Protochlorophyllide Reductase Catalyzing A Key Reduction For Greening In The Dark
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2010-04-21 Classification: OXIDOREDUCTASE Ligands: SF4, PMR |
 |
Structure Of The Light-Independent Protochlorophyllide Reductase Catalyzing A Key Reduction For Greening In The Dark
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2010-04-21 Classification: OXIDOREDUCTASE Ligands: SF4 |
 |
Structure Of The Light-Independent Protochlorophyllide Reductase Catalyzing A Key Reduction For Greening In The Dark
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2010-04-21 Classification: OXIDOREDUCTASE Ligands: SF4 |
 |
Structure Of The Light-Independent Protochlorophyllide Reductase Catalyzing A Key Reduction For Greening In The Dark
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2010-04-21 Classification: OXIDOREDUCTASE Ligands: SF4 |
 |
Structure Of The Light-Independent Protochlorophyllide Reductase Catalyzing A Key Reduction For Greening In The Dark
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2010-04-21 Classification: OXIDOREDUCTASE Ligands: SF4 |
 |
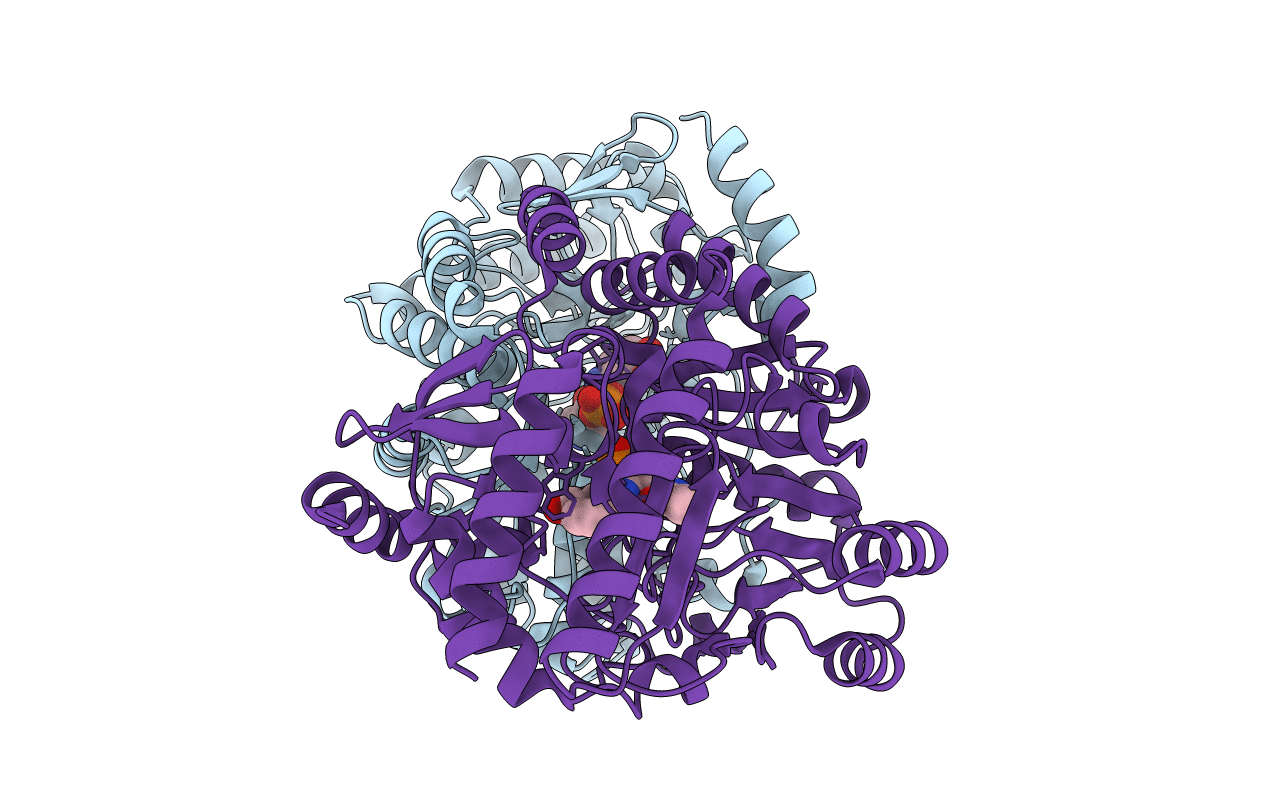
Solution Nmr Structure Of The Pcp_Red Domain Of Light-Independent Protochlorophyllide Reductase Subunit B From Chlorobium Tepidum. Northeast Structural Genomics Consortium Target Ctr69A
Organism: Chlorobaculum tepidum
Method: SOLUTION NMR Release Date: 2010-02-16 Classification: OXIDOREDUCTASE |
 |
Organism: Synechococcus elongatus pcc 6301
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2009-11-24 Classification: ISOMERASE Ligands: PXG |
 |
Organism: Synechococcus elongatus pcc 6301
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-11-24 Classification: ISOMERASE Ligands: PMP |
 |
Organism: Synechococcus elongatus pcc 6301
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2009-11-24 Classification: ISOMERASE Ligands: PMP, GAB |
 |
Crystal Structure Of The L Protein Of Rhodobacter Sphaeroides Light-Independent Protochlorophyllide Reductase (Bchl) With Mgadp Bound: A Homologue Of The Nitrogenase Fe Protein
Organism: Rhodobacter sphaeroides 2.4.1
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2009-03-24 Classification: OXIDOREDUCTASE Ligands: MG, ADP, SF4 |
 |
Structure Of Protoporphyrinogen Oxidase From Myxococcus Xanthus With Acifluorfen
Organism: Myxococcus xanthus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2006-10-17 Classification: OXIDOREDUCTASE Ligands: ACJ, FAD, GOL, TWN |
 |
Organism: Myxococcus xanthus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2006-10-17 Classification: OXIDOREDUCTASE Ligands: FAD, GOL, TWN |
 |
Glutamate-1-Semialdehyde 2,1-Aminomutase From Thermosynechococcus Elongatus
Organism: Synechococcus elongatus
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2006-03-29 Classification: ISOMERASE Ligands: PLR |
 |
Crystal Structure Of Glutamate-1-Semialdehyde Aminomutase In Complex With Gabaculine
Organism: Synechococcus sp.
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 1999-08-17 Classification: ISOMERASE Ligands: PMP, GAB |

