Search Count: 1,603
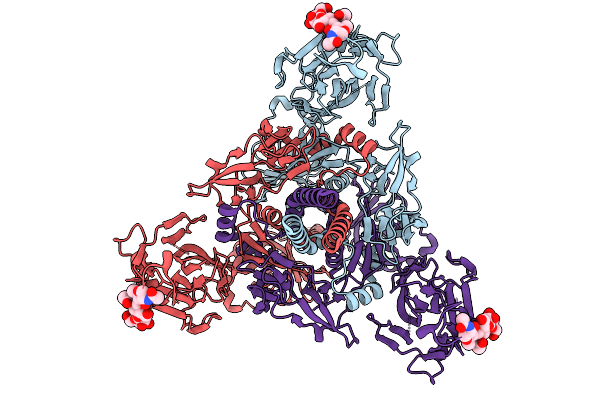 |
Organism: Human gammaherpesvirus 4
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: VIRAL PROTEIN |
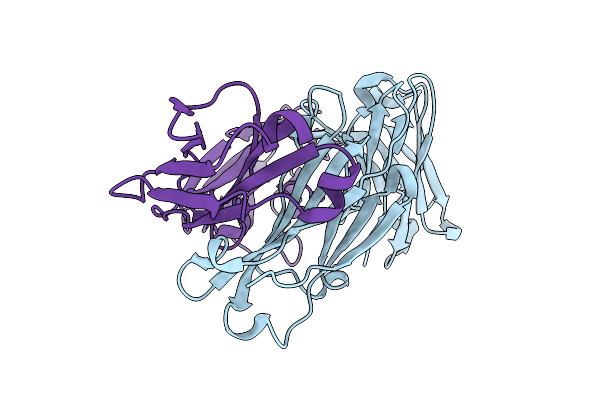 |
Crystal Structure Of Rm014, A Macaque-Derived Hiv V2-Apex-Targeting Antibody From Apexgt6 Immunization
|
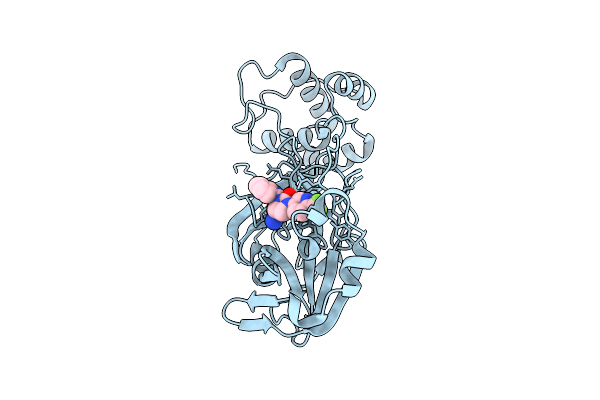 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: HYDROLASE Ligands: A1CBU |
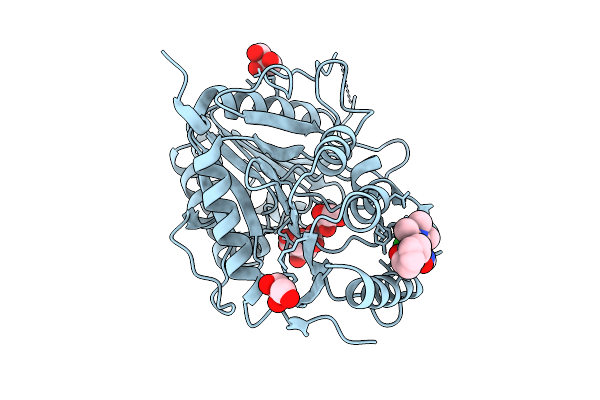 |
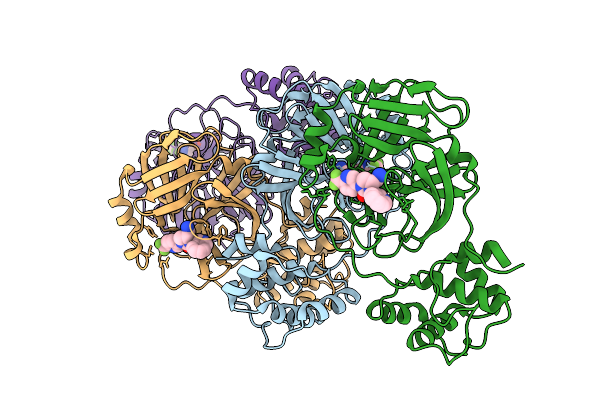
Organism: Vaccinia virus western reserve
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: VIRAL PROTEIN Ligands: GOL, CIT, A1I9D |
 |
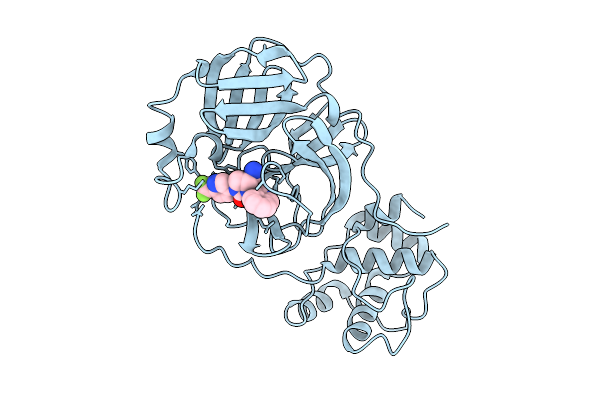
Organism: Middle east respiratory syndrome-related coronavirus
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: HYDROLASE Ligands: A1CBU |
 |
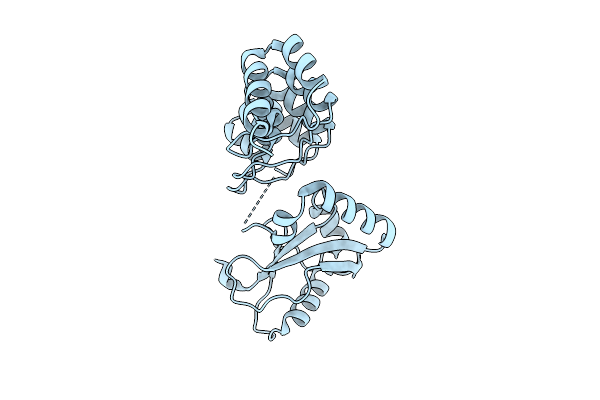
Organism: Human coronavirus oc43
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: HYDROLASE Ligands: A1CBU |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN |
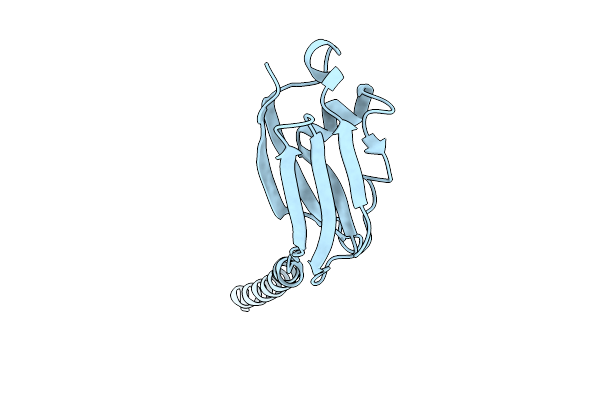 |
Organism: Natrinema sp. j7-2
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: PROTEIN FIBRIL |
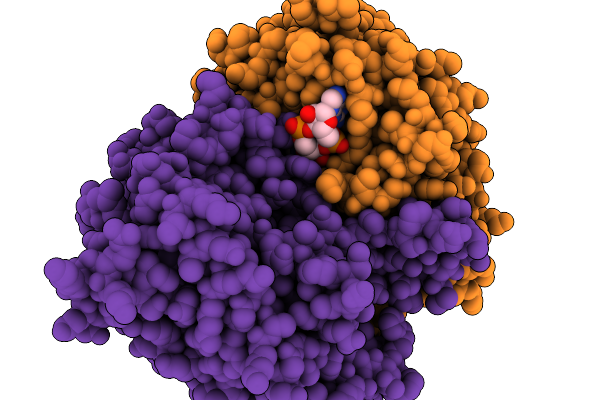 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: HYDROLASE Ligands: 4BW |
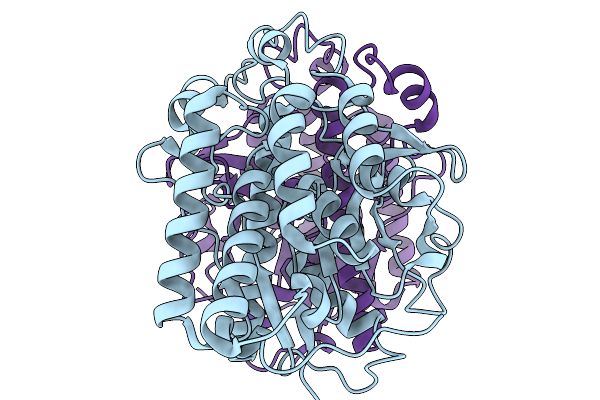 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: HYDROLASE |
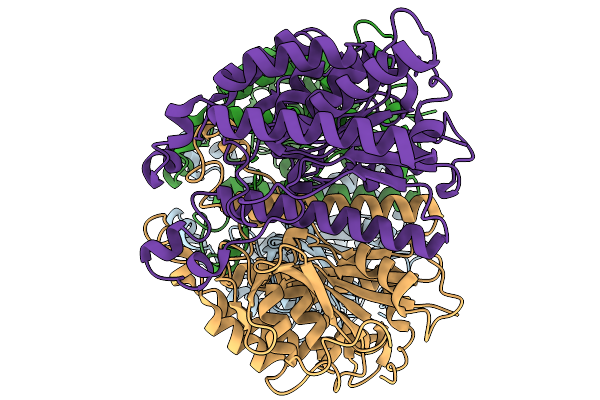 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: HYDROLASE |
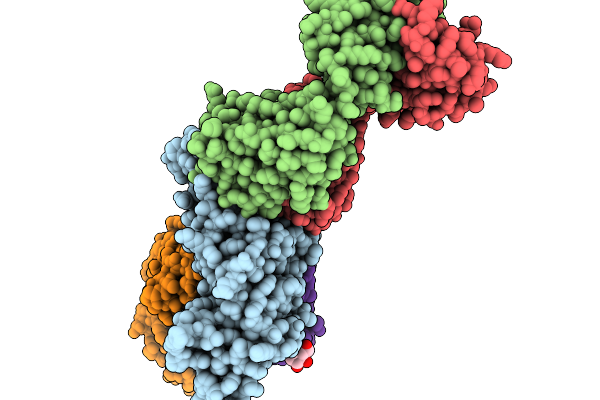 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: VIRAL PROTEIN |
 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: HYDROLASE Ligands: 4BW |
 |
Nrbf2 Coiled Coil Domain Promotes Autophagy By Strengthening Association With Vps15 In The Pi3Kc3 Complex
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: STRUCTURAL PROTEIN |
 |
Crystal Structure Of Tagatose 4-Epimerase With One Glycerol From Thermoprotei Archaeon
Organism: Thermoprotei archaeon
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: ISOMERASE Ligands: EDO, PEG, 1PG, NI, GOL |
 |
Organism: Thermoprotei archaeon
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: ISOMERASE Ligands: EDO, PEG, 1PG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: PROTEIN FIBRIL |
 |
Cryo-Em Structure Of A Tri-Heme Cytochrome-Associated Rc-Lh1 Complex From A Marine Photoheterotrophic Bacterium, Purified With Edta-Containing Solutions
Organism: Dinoroseobacter shibae dfl 12 = dsm 16493
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: PHOTOSYNTHESIS Ligands: BCL, SPN, MW9, LMT, FE, BPH, U10, CDL, HEC |

