Search Count: 6,918
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: HYDROLASE Ligands: ATP, ADP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: HYDROLASE Ligands: ATP, ADP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: HYDROLASE Ligands: ATP, ADP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: CHAPERONE Ligands: ADP |
 |
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: CHAPERONE |
 |
 |
 |
Organism: salmonella enterica subsp. enterica serovar typhi
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: PROTEIN FIBRIL Ligands: LHG |
 |
 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: CELL ADHESION |
 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: CELL ADHESION |
 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: CELL ADHESION |
 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: CELL ADHESION |
 |
A Darpin Fused To The 1Tel Crystallization Chaperone Via A Direct Helical Fusion
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: AKG, NH4, GOL, EOH, K, CL, PO4 |
 |
Organism: Brucella abortus 2308
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: STRUCTURAL PROTEIN Ligands: CL |
 |
Organism: Saccharomyces cerevisiae s288c
Method: SOLUTION NMR Release Date: 2025-12-10 Classification: CHAPERONE |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: CHAPERONE Ligands: ADP, MG |
 |
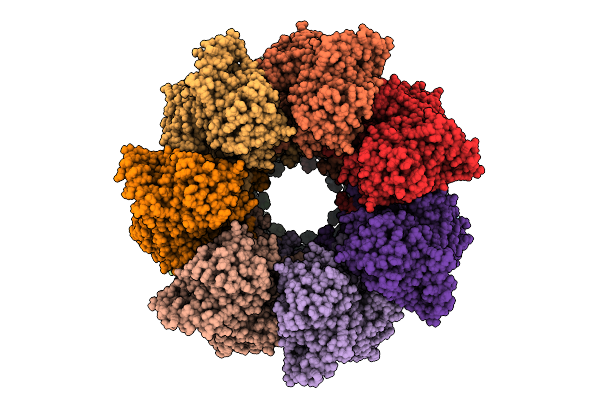
Ancestral Group Iii Chaperonin (Aciii) Double-Ring In Open Conformation Including The Equatorial And Intermediate Domains (Residues 12-200 And 356-507)
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: CHAPERONE |
 |
Ancestral Fibrobacteres-Chlorobi-Bacteroidetes Group Chaperonin (Afcb) Double-Ring In Open Conformation
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: CHAPERONE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: CHAPERONE Ligands: ATP, MG, K |

