Search Count: 8,952
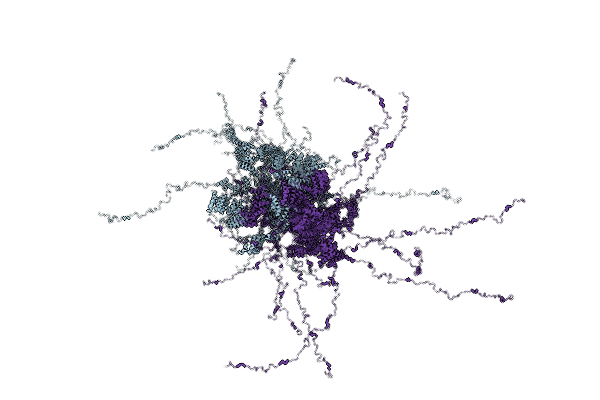 |
Organism: Saccharomyces cerevisiae s288c
Method: SOLUTION NMR Release Date: 2025-12-10 Classification: CHAPERONE |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: CHAPERONE Ligands: ADP, MG |
 |
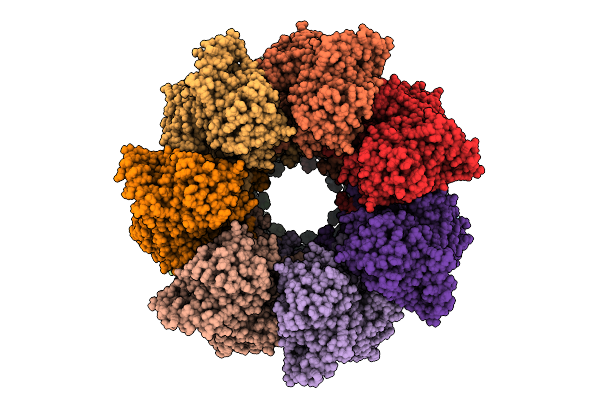
Ancestral Group Iii Chaperonin (Aciii) Double-Ring In Open Conformation Including The Equatorial And Intermediate Domains (Residues 12-200 And 356-507)
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: CHAPERONE |
 |
Ancestral Fibrobacteres-Chlorobi-Bacteroidetes Group Chaperonin (Afcb) Double-Ring In Open Conformation
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: CHAPERONE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: CHAPERONE Ligands: ATP, MG, K |
 |
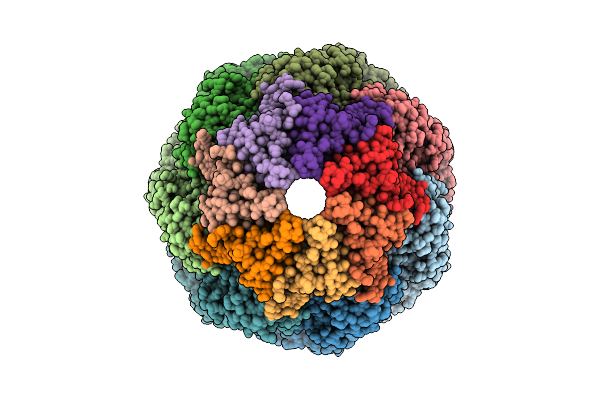
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: CHAPERONE Ligands: ATP, MG, K |
 |
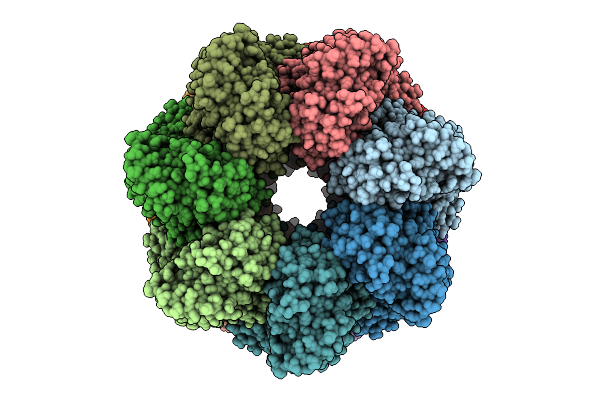
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: CHAPERONE Ligands: ATP, MG, K |
 |
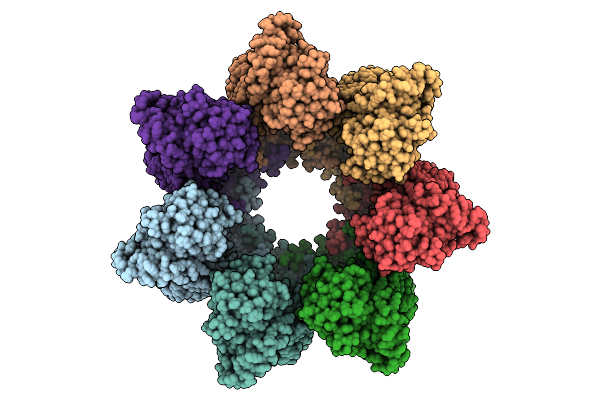
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: CHAPERONE Ligands: ATP, MG, K |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: CHAPERONE Ligands: ADP, MG |
 |
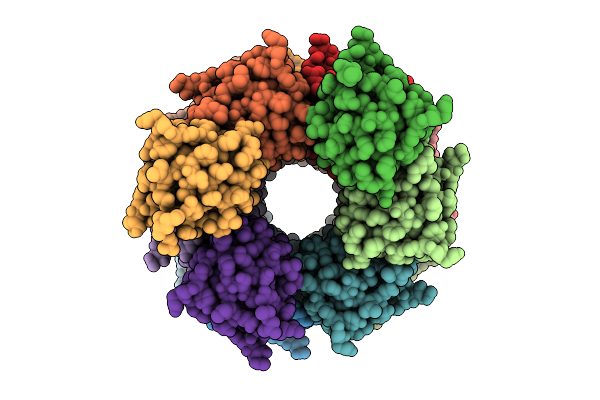
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: CHAPERONE Ligands: ATP, MG, K |
 |
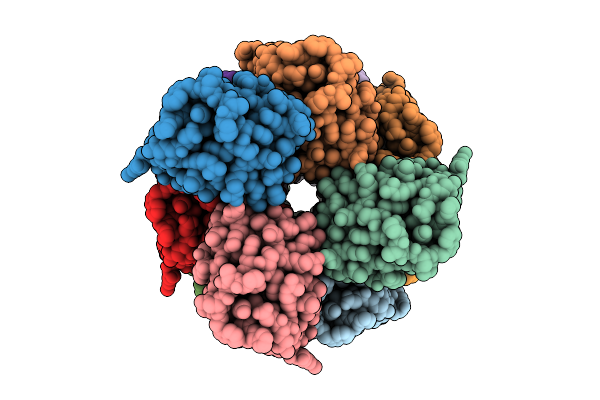
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: IMMUNE SYSTEM |
 |
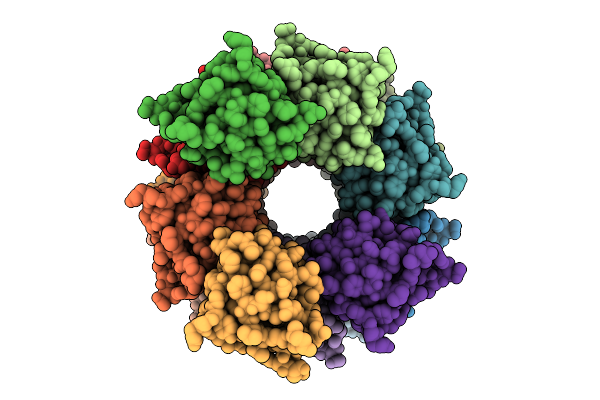
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: IMMUNE SYSTEM |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: CHAPERONE Ligands: AGS |

