Search Count: 77
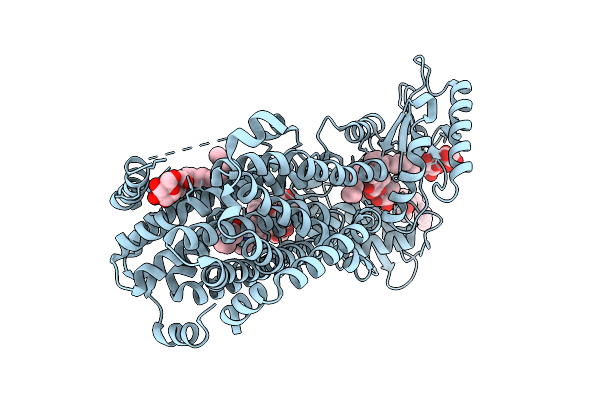 |
Crystal Structure Of Mycolic Acid Transporter Mmpl3 From Mycobacterium Smegmatis Complexed With Indolcarboxamide
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN Ligands: A1L5O, LMT |
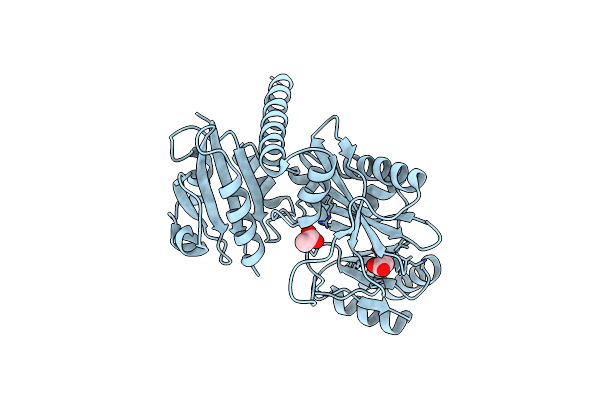 |
Crystal Structure Of Staphylococcus Aureus D-Alanine D-Alanine Ligase Enzyme In Complex With Acetate
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2024-05-08 Classification: LIGASE Ligands: ACT |
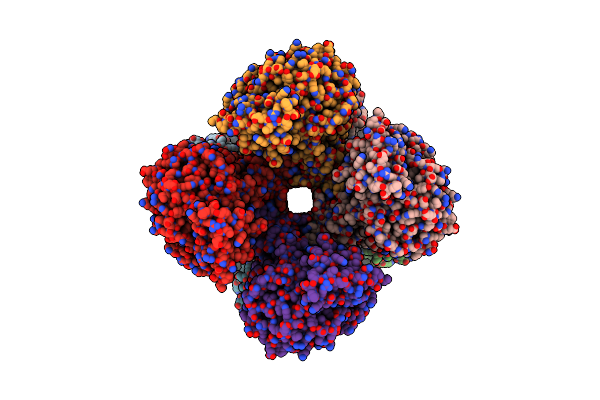 |
Cryoem Structure Of Aspergillus Nidulans Utp-Glucose-1-Phosphate Uridylyltransferase
Organism: Aspergillus nidulans fgsc a4
Method: ELECTRON MICROSCOPY Release Date: 2023-06-28 Classification: SUGAR BINDING PROTEIN |
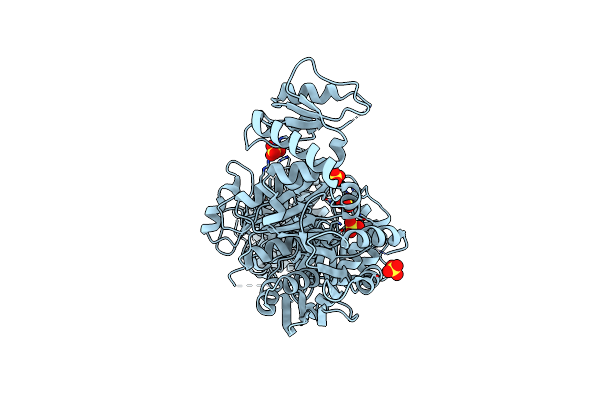 |
Architecture Of The Mure-Murf Ligase Bacterial Cell Wall Biosynthesis Complex
Organism: Bordetella pertussis 18323
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2023-06-07 Classification: LIGASE Ligands: SO4 |
 |
Crystal Structure Of Mycobacterium Thermoresistibile Mure In Complex With Adp And 2,6-Diaminopimelic Acid
Organism: Mycolicibacterium thermoresistibile
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2023-04-05 Classification: LIGASE Ligands: MG, GOL, ADP, API |
 |
Structure Of The Laspartomycin C Double Mutant G4D D-Allo-Thr9D-Dap In Complex With Geranyl Phosphate
Organism: Streptomyces viridochromogenes
Method: X-RAY DIFFRACTION Resolution:1.04 Å Release Date: 2022-02-02 Classification: ANTIBIOTIC Ligands: CA, RDZ, 9GE |
 |
Structure Of The Laspartomycin C Friulimicin-Like Mutant In Complex With Geranyl Phosphate
Organism: Streptomyces viridochromogenes
Method: X-RAY DIFFRACTION Resolution:1.14 Å Release Date: 2022-02-02 Classification: ANTIBIOTIC Ligands: CA, RDZ, CL, CD, NA |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2021-01-20 Classification: BIOSYNTHETIC PROTEIN Ligands: EDO, T4H |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-01-13 Classification: BIOSYNTHETIC PROTEIN Ligands: CIT, SYQ, T3Z, IPA, DMS |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2021-01-13 Classification: BIOSYNTHETIC PROTEIN Ligands: DMS, NUJ |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2020-12-23 Classification: BIOSYNTHETIC PROTEIN Ligands: O3D, DMS, IPA |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2020-12-23 Classification: BIOSYNTHETIC PROTEIN Ligands: LVV, DMS |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2020-12-23 Classification: BIOSYNTHETIC PROTEIN Ligands: CIT, SZE, IPA |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2020-12-23 Classification: BIOSYNTHETIC PROTEIN Ligands: EDO, SZB, IPA |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2020-12-23 Classification: BIOSYNTHETIC PROTEIN Ligands: DMS, IPA, SZ2, CIT |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2020-12-23 Classification: BIOSYNTHETIC PROTEIN Ligands: IPA, SZK |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2020-12-23 Classification: BIOSYNTHETIC PROTEIN Ligands: EDO, SZN |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2020-12-23 Classification: BIOSYNTHETIC PROTEIN Ligands: EDO, SYZ |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2020-12-23 Classification: BIOSYNTHETIC PROTEIN Ligands: IPA |
 |
Crystal Structure Of E.Coli Mure - C269S C340S C450S In Complex With Ellman'S Reagent
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2020-12-23 Classification: BIOSYNTHETIC PROTEIN Ligands: 9JT, IPA, CIT, EDO |

