Search Count: 38
 |
Sirt2 Structure In Complex With H3K18Myr Peptide And Native Nad: Pre-Catalysis State 3
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: HYDROLASE Ligands: PEG, EDO, NAD, GOL, ZN, MYR |
 |
Sirt2 Structure In Complex With H3K18Myr Peptide And Native Nad: Pre-Catalysis State 1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: HYDROLASE Ligands: NAD, PEG, EDO, GOL, ZN, MYR |
 |
Sirt2 Structure In Complex With H3K18Myr Peptide And Native Nad: Pre-Catalysis State 2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: HYDROLASE Ligands: NAD, EDO, GOL, ZN, MYR |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: HYDROLASE Ligands: EDO, ZN, MYR |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: HYDROLASE Ligands: EDO, ZN, MYR |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: HYDROLASE Ligands: NCA, GOL, ZN, YDD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: HYDROLASE Ligands: NAD, EDO, PEG, ZN, MYR |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: HYDROLASE Ligands: NAD, ZN, MYR |
 |
Cryo-Em Structure Of The Mcm-Orc (Mo) Complex Featuring An Orc2 Regulatory Domain Involved In Cell Cycle Regulation Of Mcm-Dh Loading For Dna Replication.
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: REPLICATION Ligands: ATP, MG, ADP, ZN |
 |
Discovery Of (4-Pyrazolyl)-2-Aminopyrimidines As Potent And Selective Inhibitors Of Cyclin-Dependent Kinase 2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2024-02-14 Classification: TRANSFERASE/INHIBITOR Ligands: XKU |
 |
Structure Of The Ddb1-Ambra1 E3 Ligase Receptor Complex Linked To Cell Cycle Regulation
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: STRUCTURAL PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-11-01 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-11-01 Classification: SIGNALING PROTEIN Ligands: FE, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-10-25 Classification: SIGNALING PROTEIN Ligands: FE, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-10-25 Classification: SIGNALING PROTEIN,HYDROLASE Ligands: FE, ZN |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2020-10-28 Classification: RIBOSOME Ligands: MG, ZN, ADP, ATP, SF4 |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2020-10-07 Classification: RIBOSOME Ligands: ZN, ADP, MG, ATP, SF4 |
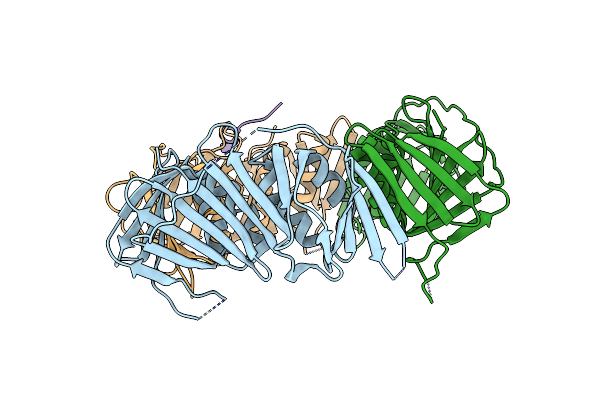 |
The Structure Of Schizosaccharomyces Pombe Pcna In Complex With An Spd1 Derived Peptide
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2020-02-05 Classification: DNA BINDING PROTEIN |
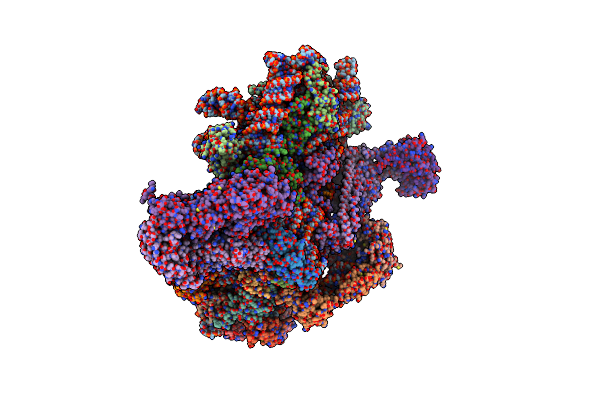 |
Structure Of A Partial Yeast 48S Preinitiation Complex In Open Conformation.
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Resolution:5.15 Å Release Date: 2019-07-31 Classification: RIBOSOME Ligands: 7NO, MG, ZN, GCP |
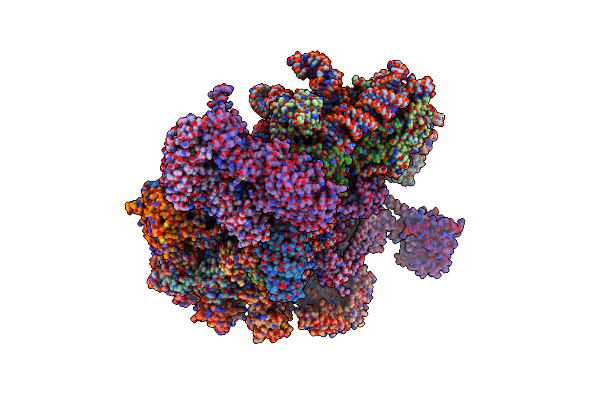 |
Structure Of A Partial Yeast 48S Preinitiation Complex In Closed Conformation
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Kluyveromyces lactis (strain atcc 8585 / cbs 2359 / dsm 70799 / nbrc 1267 / nrrl y-1140 / wm37)
Method: ELECTRON MICROSCOPY Release Date: 2019-06-26 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |

