Search Count: 27
 |
Structure Of S. Cerevisiae Rad24-Rfc Loading The 9-1-1 Clamp Onto A 10-Nt Gapped Dna In Step 1 (Open 9-1-1 And Shoulder Bound Dna Only)
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-06-14 Classification: CELL CYCLE/DNA Ligands: MG, AGS, ADP |
 |
Structure Of S. Cerevisiae Rad24-Rfc Loading The 9-1-1 Clamp Onto A 10-Nt Gapped Dna In Step 2 (Open 9-1-1 Ring And Flexibly Bound Chamber Dna)
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-06-14 Classification: CELL CYCLE/DNA Ligands: AGS, MG, ADP |
 |
Structure Of S. Cerevisiae Rad24-Rfc Loading The 9-1-1 Clamp Onto A 10-Nt Gapped Dna In Step 3 (Open 9-1-1 And Stably Bound Chamber Dna)
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-06-14 Classification: CELL CYCLE/DNA Ligands: AGS, MG, ADP |
 |
Structure Of S. Cerevisiae Rad24-Rfc Loading The 9-1-1 Clamp Onto A 10-Nt Gapped Dna In Step 4 (Partially Closed 9-1-1 And Stably Bound Chamber Dna)
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-06-14 Classification: CELL CYCLE/DNA Ligands: AGS, MG, ADP |
 |
Structure Of S. Cerevisiae Rad24-Rfc Loading The 9-1-1 Clamp Onto A 10-Nt Gapped Dna In Step 5 (Closed 9-1-1 And Stably Bound Chamber Dna)
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-06-14 Classification: CELL CYCLE/DNA Ligands: MG, AGS, ADP |
 |
Structure Of S. Cerevisiae Rad24-Rfc Loading The 9-1-1 Clamp Onto A 5-Nt Gapped Dna (9-1-1 Encircling Fully Bound Dna)
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-06-14 Classification: CELL CYCLE/DNA Ligands: AGS, MG, ADP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-02-08 Classification: CELL CYCLE/DNA Ligands: ZN, MG, ATP, ADP |
 |
The Cryo-Em Structure Of Cenp-A Nucleosome In Complex With Cenp-C Peptide And Cenp-N N-Terminal Domain
Organism: Gallus gallus, Homo sapiens, Unidentified cloning vector, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-02-10 Classification: CELL CYCLE/DNA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-05-20 Classification: CELL CYCLE/DNA Ligands: ANP |
 |
Crystal Structure Of Human Smc1-Smc3 Hinge Domain Heterodimer In North-Open Conformation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.54 Å Release Date: 2020-05-20 Classification: CELL CYCLE/DNA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-05-20 Classification: CELL CYCLE/DNA Ligands: ANP, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2018-09-19 Classification: CELL CYCLE/DNA Binding Ligands: SO4 |
 |
Structures Of The No Factor Slma Bound To Dna And The Cytoskeletal Cell Division Protein Ftsz
Organism: Vibrio cholerae serotype o1 (strain atcc 39315 / el tor inaba n16961), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2016-04-13 Classification: CELL CYCLE/DNA |
 |
Structure Of The E. Coli Nucleoid Occlusion Protein Slma Bound To Dna And The C-Terminal Tail Of The Cytoskeletal Cell Division Protein Ftsz
Organism: Escherichia coli (strain k12), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-04-13 Classification: CELL CYCLE/DNA |
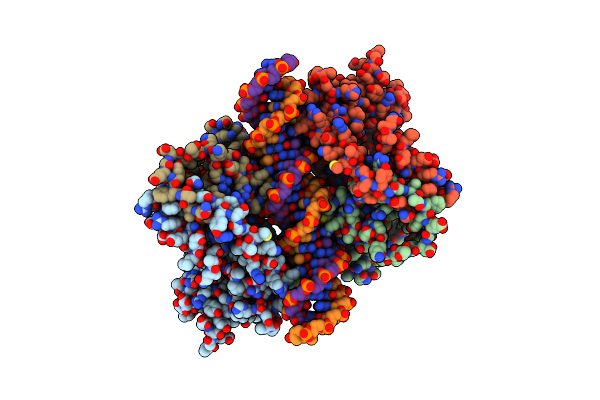 |
An Induced Fit Mechanism Regulates P53 Dna Binding Kinetics To Confer Sequence Specificity
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2011-05-11 Classification: CELL CYCLE/DNA Ligands: ZN |
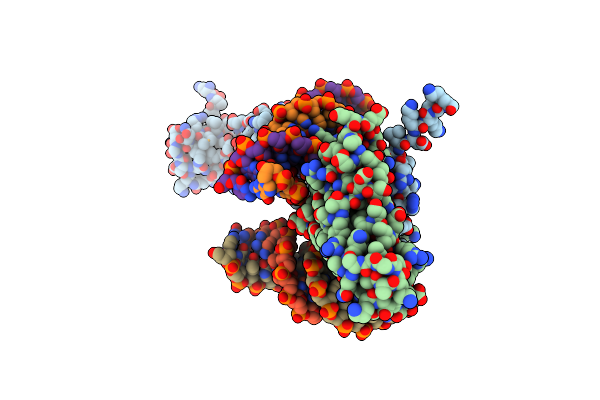 |
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2007-02-20 Classification: CELL CYCLE/DNA |
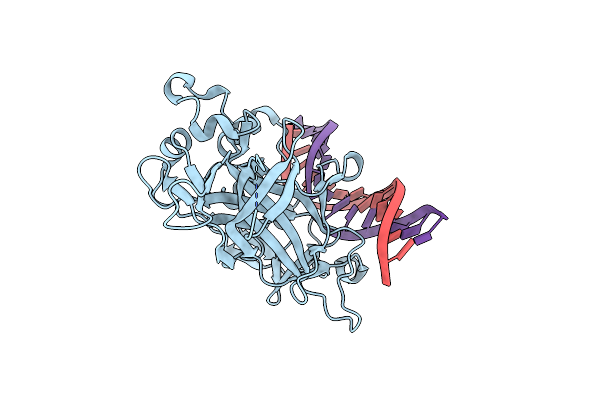 |
Principles Of Protein-Dna Recognition Revealed In The Structural Analysis Of Ndt80-Mse Dna Complexes
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2006-03-21 Classification: CELL CYCLE/DNA |
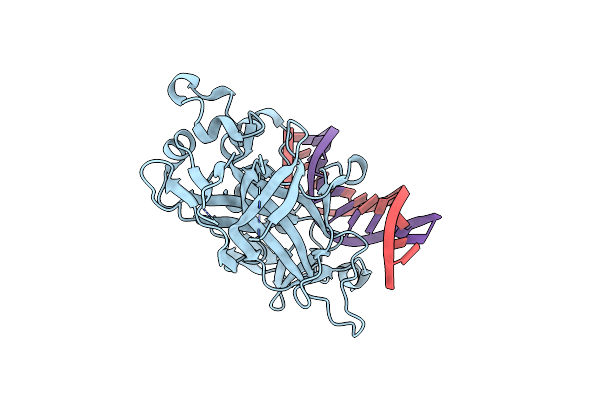 |
Principles Of Protein-Dna Recognition Revealed In The Structural Analysis Of Ndt80-Mse Dna Complexes
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2006-03-21 Classification: CELL CYCLE/DNA |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2006-03-21 Classification: CELL CYCLE/DNA |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2006-03-21 Classification: CELL CYCLE/DNA |

