Search Count: 9
 |
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2014-02-05 Classification: FLUORESCENT PROTEIN |
 |
1.10A Resolution Crystal Structure Of A Superfolder Green Fluorescent Protein (W57A) Mutant
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2013-12-18 Classification: FLUORESCENT PROTEIN Ligands: EDO |
 |
1.60A Resolution Crystal Structure Of A Superfolder Green Fluorescent Protein (W57G) Mutant
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2013-12-18 Classification: FLUORESCENT PROTEIN |
 |
Crystal Structure Of Unliganded Escherichia Coli Dihydrofolate Reductase. Ligand-Induced Conformational Changes And Cooperativity In Binding
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1994-01-31 Classification: OXIDOREDUCTASE Ligands: CL, MTX, CA |
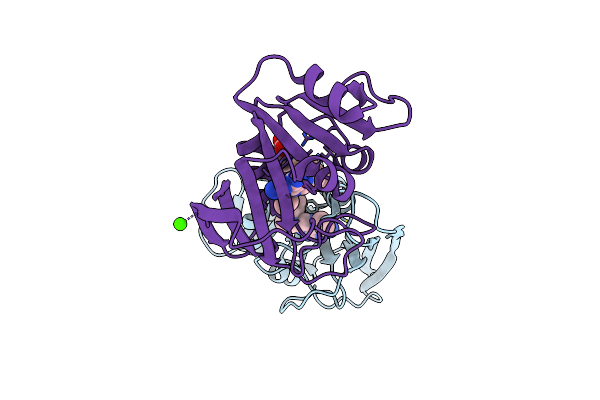 |
Crystal Structure Of Unliganded Escherichia Coli Dihydrofolate Reductase. Ligand-Induced Conformational Changes And Cooperativity In Binding
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 1994-01-31 Classification: OXIDOREDUCTASE Ligands: CL, MTX, EOH, CA |
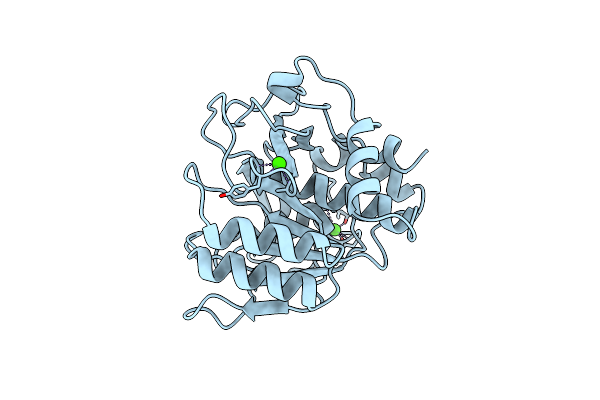 |
The Crystal Structure Of The Bacillus Lentus Alkaline Protease, Subtilisin Bl, At 1.4 Angstroms Resolution
Organism: Bacillus lentus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 1994-01-31 Classification: SERINE PROTEASE Ligands: CA |
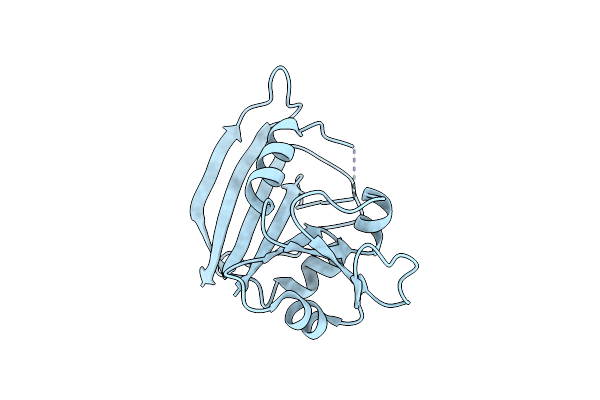 |
Crystal Structure Of Unliganded Escherichia Coli Dihydrofolate Reductase. Ligand-Induced Conformational Changes And Cooperativity In Binding
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 1990-07-15 Classification: OXIDOREDUCTASE Ligands: CL |
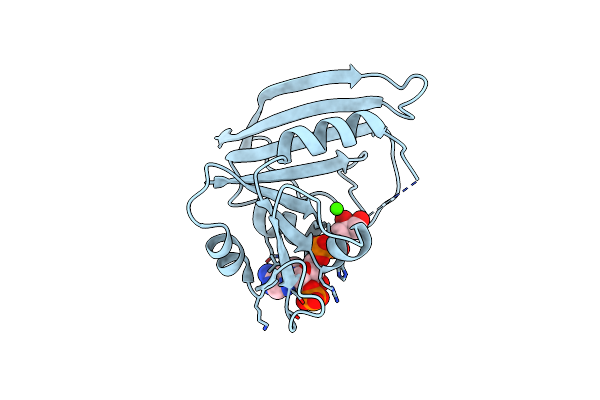 |
Crystal Structures Of Escherichia Coli Dihydrofolate Reductase. The Nadp+ Holoenzyme And The Folate(Dot)Nadp+ Ternary Complex. Substrate Binding And A Model For The Transition State
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1990-07-15 Classification: OXIDOREDUCTASE Ligands: CA, NAP |
 |
Crystal Structures Of Escherichia Coli Dihydrofolate Reductase. The Nadp+ Holoenzyme And The Folate(Dot)Nadp+ Ternary Complex. Substrate Binding And A Model For The Transition State
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 1990-07-15 Classification: OXIDOREDUCTASE Ligands: FOL, NAP |

