Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
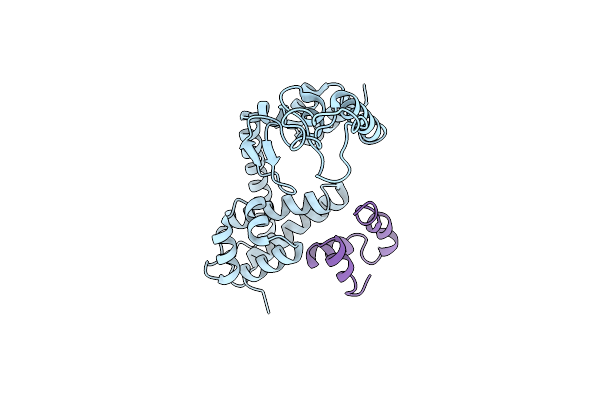 |
Organism: Staphylococcus aureus, Staphylococcus phage 80alpha
Method: X-RAY DIFFRACTION Resolution:2.97 Å Release Date: 2022-09-28 Classification: GENE REGULATION |
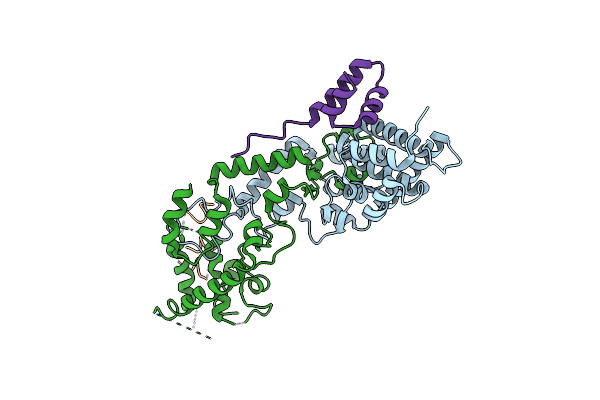 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2022-07-27 Classification: DNA BINDING PROTEIN |
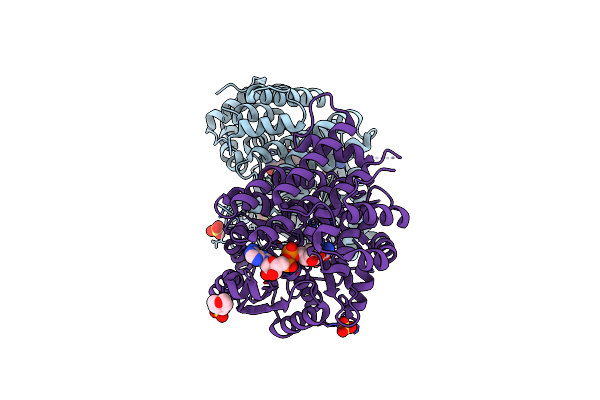 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2020-08-26 Classification: OXIDOREDUCTASE Ligands: NAD, SO4, EDO, FE, MES |
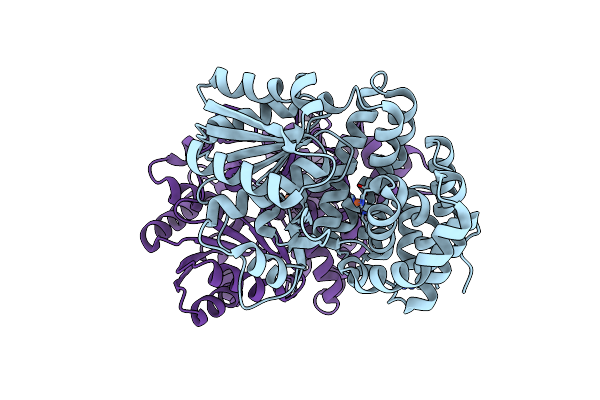 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2020-08-26 Classification: OXIDOREDUCTASE Ligands: FE |
 |
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2020-06-24 Classification: OXIDOREDUCTASE Ligands: NAD, ZN |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:4.36 Å Release Date: 2020-04-15 Classification: CELL CYCLE |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:3.47 Å Release Date: 2020-04-01 Classification: CELL CYCLE |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2020-04-01 Classification: CELL CYCLE |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2020-04-01 Classification: CELL CYCLE |
 |
Cryo-Em Structure Of Aldehyde-Alcohol Dehydrogenase Reveals A High-Order Helical Architecture Critical For Its Activity
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2019-08-21 Classification: HYDROLASE |
 |
Organism: Feline calicivirus
Method: ELECTRON MICROSCOPY Release Date: 2019-01-16 Classification: VIRUS Ligands: K |
 |
Feline Calicivirus Strain F9 Bound To A Soluble Ectodomain Fragment Of Feline Junctional Adhesion Molecule A - Leading To Assembly Of A Portal Structure At A Unique Three-Fold Axis.
Organism: Felis catus, Feline calicivirus strain f9
Method: ELECTRON MICROSCOPY Release Date: 2019-01-16 Classification: VIRUS Ligands: K |
 |
The Crystal Structure Of The Ferredoxin Receptor Fusa From Pectobacterium Atrosepticum Scri1043
Organism: Pectobacterium atrosepticum
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2016-08-31 Classification: TRANSPORT PROTEIN Ligands: LDA, BOG |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2016-08-31 Classification: ELECTRON TRANSPORT Ligands: FES, CL |
 |
Organism: Solanum tuberosum
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2016-08-31 Classification: ELECTRON TRANSPORT Ligands: FES |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2016-07-20 Classification: HYDROLASE |
 |
Organism: Human gut metagenome
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2015-11-18 Classification: TRANSPORT PROTEIN Ligands: PO4 |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-08-05 Classification: HYDROLASE |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2015-08-05 Classification: HYDROLASE Ligands: CIT, NI |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.65 Å Release Date: 2015-07-15 Classification: LIPID BINDING PROTEIN |