Search Count: 29
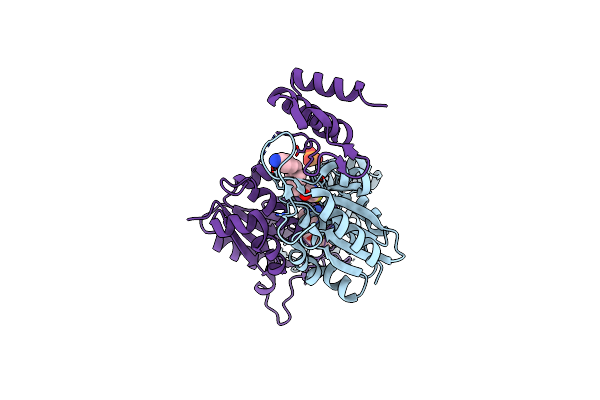 |
Crystal Structure Of Trmd From Pseudomonas Aeruginosa In Complex With Active-Site Inhibitor
Organism: Pseudomonas aeruginosa ucbpp-pa14
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2019-09-04 Classification: TRANSFERASE Ligands: BWR, PO4, SAM |
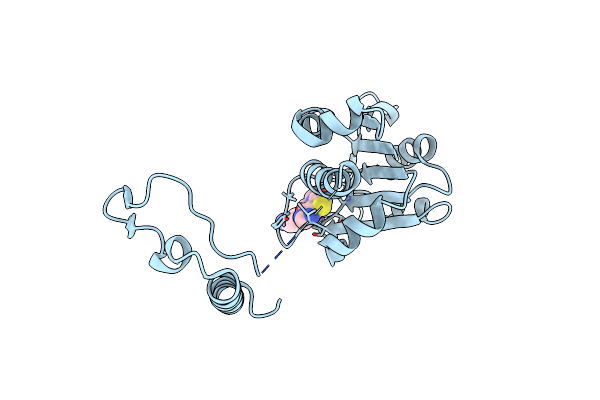 |
Crystal Structure Of Trmd From Mycobacterium Tuberculosis In Complex With Active-Site Inhibitor
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-09-04 Classification: TRANSFERASE/INHIBITOR Ligands: BWR |
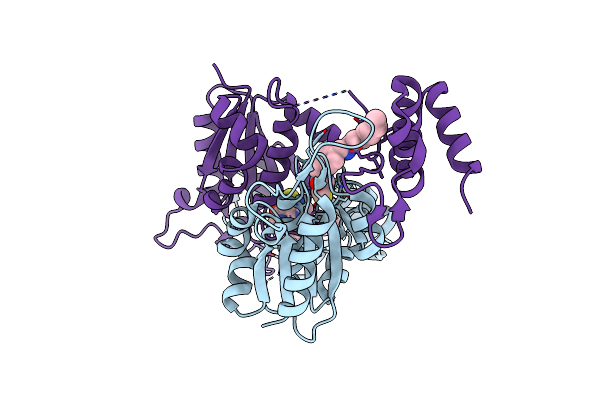 |
Crystal Structure Of Trmd From Pseudomonas Aeruginosa In Complex With Active-Site Inhibitor
Organism: Pseudomonas aeruginosa ucbpp-pa14
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2019-03-27 Classification: TRANSFERASE Ligands: 9D0 |
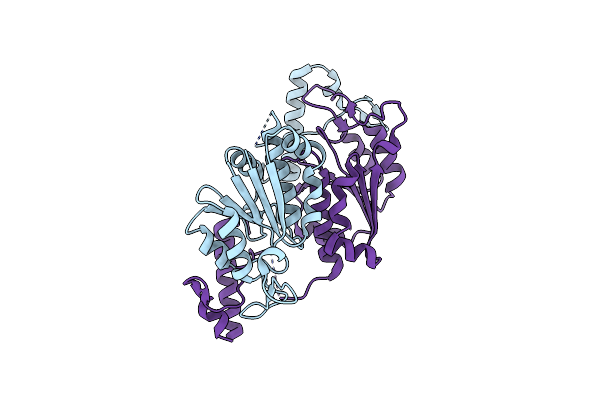 |
Organism: Mycobacterium tuberculosis (strain cdc 1551 / oshkosh)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-03-06 Classification: TRANSFERASE |
 |
Crystal Structure Of Trmd From Mycobacterium Tuberculosis In Complex With S-Adenosyl Homocysteine (Sah)
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-03-06 Classification: TRANSFERASE Ligands: SAH, GOL |
 |
Crystal Structure Of Trmd From Mycobacterium Tuberculosis In Complex With Active-Site Inhibitor
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-03-06 Classification: TRANSFERASE Ligands: 9D6 |
 |
Crystal Structure Of Trmd From Mycobacterium Tuberculosis In Complex With Active-Site Inhibitor
Organism: Mycobacterium tuberculosis cdc 1551
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2019-03-06 Classification: TRANSFERASE Ligands: 9D0 |
 |
Crystal Structure Of Trmd From Pseudomonas Aeruginosa In Complex With Active-Site Inhibitor
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2019-03-06 Classification: TRANSFERASE Ligands: 9D3, SAM |
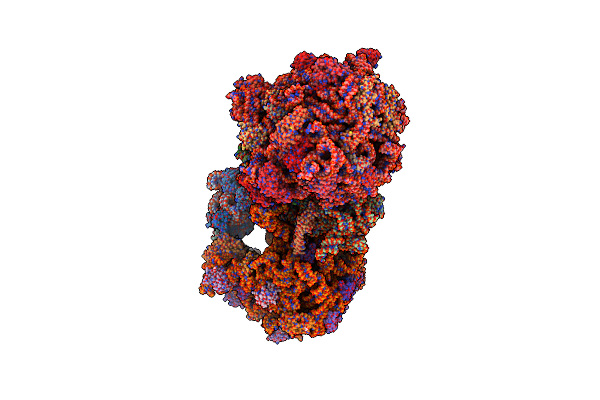 |
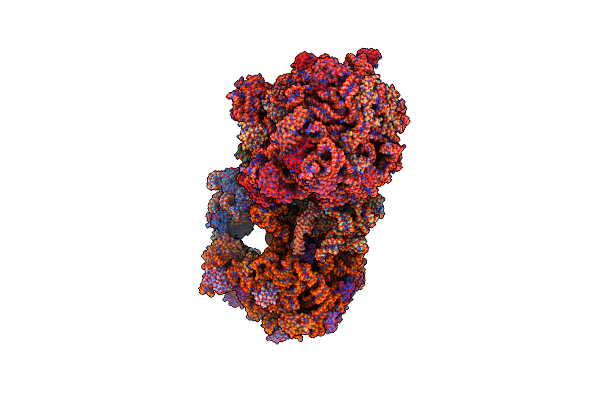 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-07-27 Classification: RIBOSOME/ANTIBIOTIC Ligands: MG, PG4, MPD, PUT, TAC, ZN, PEG, EDO, PGE, SPD, 1PE, ACY, GUN, TRS |
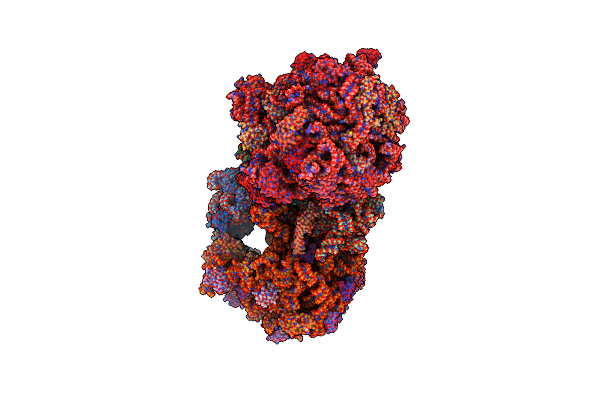 |
Structure Of The 70S E Coli Ribosome With The U1052G Mutation In The 16S Rrna Bound To Tetracycline
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2016-07-27 Classification: RIBOSOME/ANTIBIOTIC Ligands: MG, PG4, MPD, PUT, TAC, ZN, PEG, EDO, PGE, SPD, 1PE, ACY, GUN, TRS |
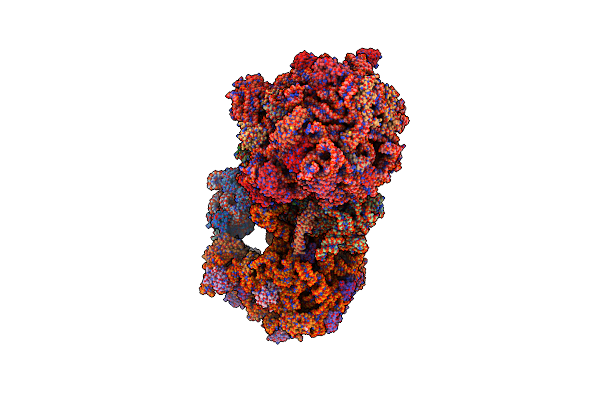 |
Organism: Escherichia coli, Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:3.32 Å Release Date: 2016-07-06 Classification: ribosome/antibiotic Ligands: MG, PG4, MPD, PUT, ZN, PEG, EDO, PGE, SPD, 1PE, ACY, GUN, TRS |
 |
Structure Of The E Coli 70S Ribosome With The U1052G Mutation In 16S Rrna Bound To Tigecycline
Organism: Escherichia coli, Escherichia coli (strain k12), Escherichia coli o139:h28 (strain e24377a / etec)
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2016-07-06 Classification: ribosome/antibiotic Ligands: MG, PG4, MPD, PUT, T1C, ZN, PEG, EDO, PGE, SPD, 1PE, ACY, GUN, TRS |
 |
Organism: Escherichia coli, Escherichia coli k-12, Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.96 Å Release Date: 2016-07-06 Classification: ribosome/antibiotic Ligands: MG, PG4, MPD, PUT, T1C, ZN, PEG, EDO, PGE, SPD, 1PE, ACY, GUN, TRS |
 |
Structure Of The Escherichia Coli Ribosome With The U1052G Mutation In The 16S Rrna
|
 |
Organism: Escherichia coli o139:h28 (strain e24377a / etec), Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:3.57 Å Release Date: 2015-03-11 Classification: transcription/transcription inhibitor Ligands: MG, ZN, 4C6 |
 |
Escherichia Coli Rna Polymerase In Complex With Squaramide Compound 14 (N-[3,4-Dioxo-2-(4-{[4-(Trifluoromethyl)Benzyl]Amino}Piperidin-1-Yl)Cyclobut-1-En-1-Yl]-3,5-Dimethyl-1,2-Oxazole-4-Sulfonamide)
Organism: Escherichia coli o139:h28 (strain e24377a / etec), Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:3.82 Å Release Date: 2015-03-11 Classification: transcription/transcription inhibitor Ligands: MG, ZN, 4C2 |
 |
Organism: Escherichia coli o139:h28 (strain e24377a / etec), Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:3.84 Å Release Date: 2015-03-11 Classification: transcription/transcription inhibitor Ligands: MG, ZN, 4C4 |
 |
Organism: Escherichia coli str. k-12 substr. mg1655, Escherichia coli, Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.09 Å Release Date: 2014-11-05 Classification: Ribosome/Antibiotic Ligands: MG, ZN, NEG |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2014-06-25 Classification: LIGASE |

