Search Count: 30
 |
Apex Domain Deletion Mutant Of Bacteriophage P2 Central Spike Protein, Membrane-Piercing Module
Organism: Bacteriophage p2
Method: X-RAY DIFFRACTION Resolution:0.90 Å Release Date: 2024-08-07 Classification: VIRAL PROTEIN Ligands: PTY |
 |
Double Alanine Apex Domain Mutant Of Bacteriophage P2 Central Spike Protein, Membrane-Piercing Module
Organism: Bacteriophage p2
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2024-08-07 Classification: VIRAL PROTEIN Ligands: PTY |
 |
Double Phenylalanine Apex Domain Mutant Of Bacteriophage P2 Central Spike Protein, Membrane-Piercing Module
Organism: Bacteriophage p2
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2024-08-07 Classification: VIRAL PROTEIN Ligands: PTY |
 |
Bacteriophage Phikz Gp163.1 Paar Repeat Protein In Complex With The C-Terminal Part Of The T4 Gp5 Beta-Helical Domain
Organism: Enterobacteria phage t4, Pseudomonas phage phikz
Method: X-RAY DIFFRACTION Release Date: 2020-05-27 Classification: viral protein, hydrolase Ligands: MG, PLM, STE, ELA, ZN |
 |
Bacteriophage Phikz Gp163.1 Paar Repeat Protein In Complex With A T4 Gp5 Beta-Helix Fragment Modified To Mimic The Phikz Central Spike Gp164
Organism: Enterobacteria phage t4, Pseudomonas phage phikz
Method: X-RAY DIFFRACTION Release Date: 2020-05-27 Classification: VIRAL PROTEIN Ligands: MG, STE, ELA, EDO, PLM, ZN |
 |
Photorhabdus Virulence Cassette (Pvc) Paar Repeat Protein Pvc10 In Complex With A T4 Gp5 Beta-Helix Fragment Modified To Mimic Pvc8, The Central Spike Protein Of Pvc
Organism: Enterobacteria phage t4, Photorhabdus luminescens subsp. laumondii (strain dsm 15139 / cip 105565 / tt01)
Method: X-RAY DIFFRACTION Release Date: 2020-05-27 Classification: VIRAL PROTEIN Ligands: MG, STE, ELA, PLM |
 |
Chimera Of Bacteriophage Obp Gp146 Central Spike Protein And A T4 Gp5 Beta-Helix Fragment
Organism: Enterobacteria phage t4, Pseudomonas phage obp
Method: X-RAY DIFFRACTION Release Date: 2020-05-27 Classification: VIRAL PROTEIN Ligands: FE2, STE, ELA, PLM, MG |
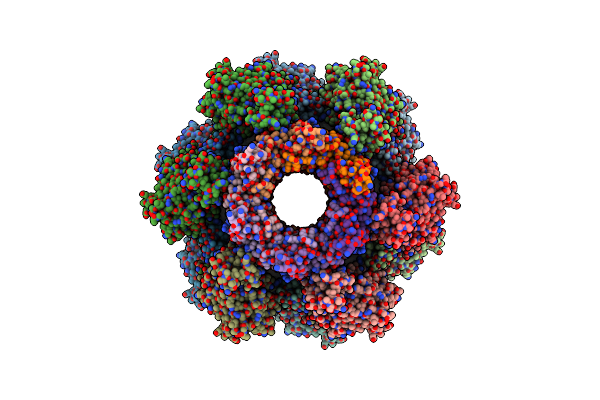 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: ANTIMICROBIAL PROTEIN |
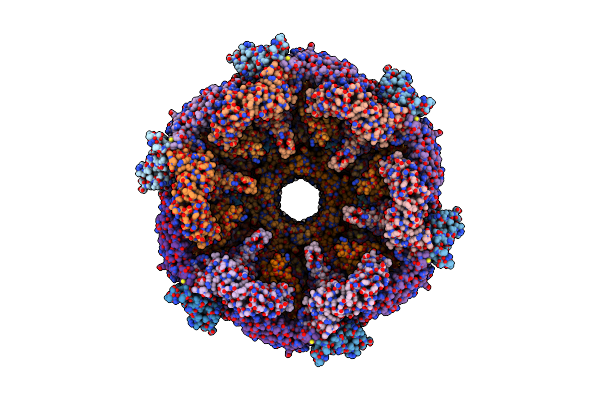 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: ANTIMICROBIAL PROTEIN |
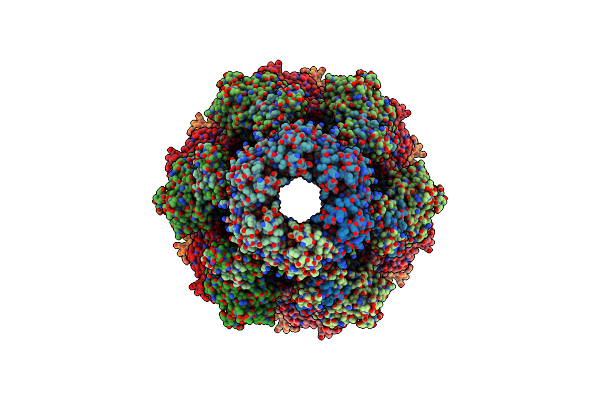 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: ANTIMICROBIAL PROTEIN |
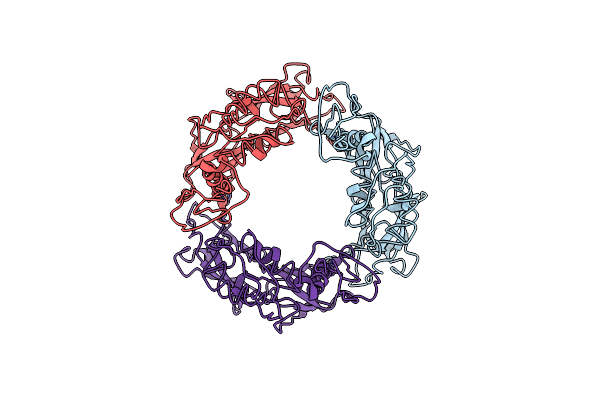 |
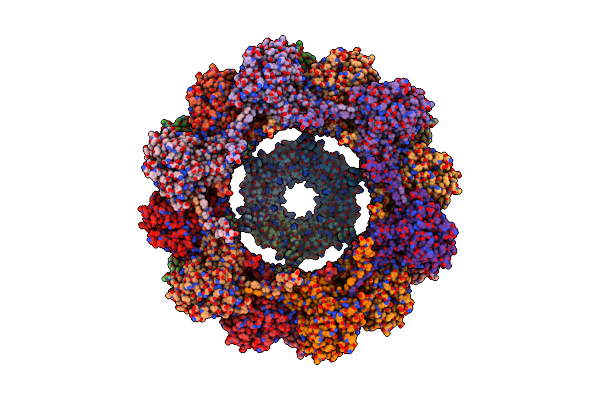
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: UNKNOWN FUNCTION |
 |
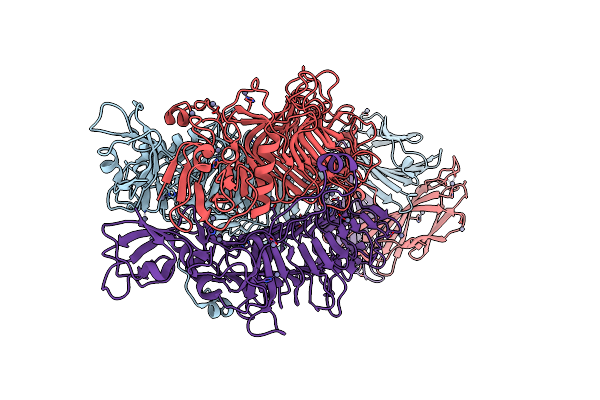
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: ANTIMICROBIAL PROTEIN |
 |
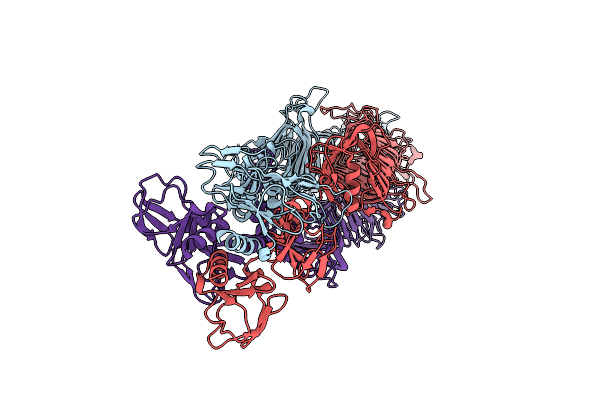
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: ANTIMICROBIAL PROTEIN |
 |
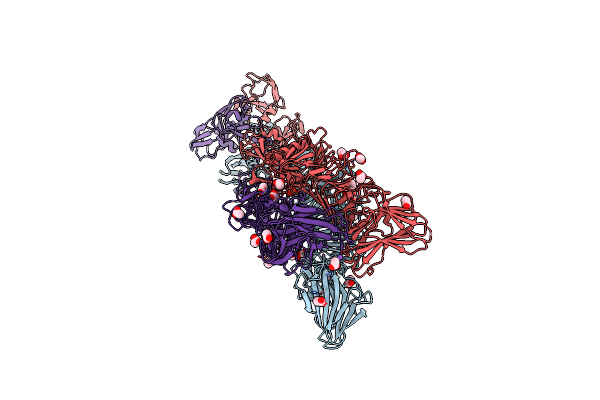
Crystal Structure Of A Tailspike Protein Gp49 From Acinetobacter Baumannii Bacteriophage Fri1, A Capsular Polysaccharide Depolymerase
Organism: Acinetobacter phage fri1
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2019-01-23 Classification: HYDROLASE Ligands: ZN |
 |
Crystal Structure Of Bacteriophage Cba120 Tailspike Protein 3 (Tsp3, Orf212)
Organism: Escherichia phage cba120
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2018-10-24 Classification: viral protein, hydrolase Ligands: CL |
 |
Crystal Structure Of Bacteriophage Cba120 Tailspike Protein 4 Enzymatically Active Domain (Tsp4Dn, Orf213)
Organism: Escherichia virus cba120
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2018-10-24 Classification: viral protein, hydrolase Ligands: ACT, K, NA, ZN, SO4, CL |
 |
Crystal Structure Of Bacteriophage Cba120 Tailspike Protein 2 Enzymatically Active Domain (Tsp2Dn, Orf211)
Organism: Escherichia phage cba120
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2018-10-24 Classification: viral protein, hydrolase Ligands: ZN, K, EDO |
 |
Crystal Structure Of Bacteriophage Cba120 Tailspike Protein 2 Enzymatically Active Domain (Tsp2Dn, Orf211) Complex With Escherichia Coli O157-Antigen
Organism: Escherichia phage cba120
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2018-10-24 Classification: viral protein, hydrolase Ligands: PGE, PG4, SO4, EDO, NA, K, CL |
 |
Structure Of P. Aeruginosa R1 Pyocin Fiber Pales_06171 Comprising C-Terminal Residues 323-701
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2018-06-27 Classification: VIRAL PROTEIN Ligands: FE, MG, NA |
 |
Structure Of P. Aeruginosa R2 Pyocin Fiber Pa0620 Comprising C-Terminal Residues 323-691
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-06-27 Classification: VIRAL PROTEIN Ligands: FE, MG, CA, EDO |

