Search Count: 16
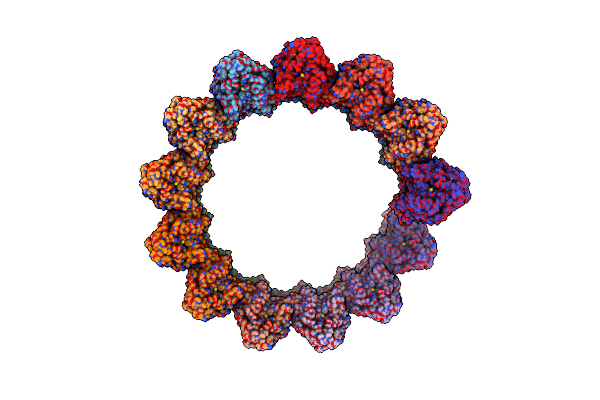 |
Structure Of The Native Microtubule Lattice Nucleated From The Yeast Spindle Pole Body
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-04-24 Classification: CELL CYCLE |
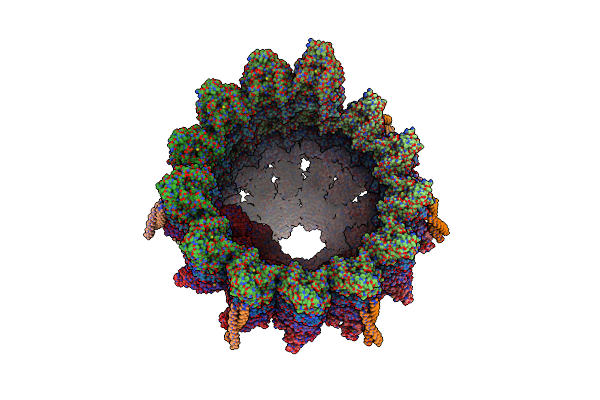 |
Structure Of The Native Y-Tubulin Ring Complex (Yturc) Capping Microtubule Minus Ends At The Spindle Pole Body
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-04-24 Classification: CELL CYCLE Ligands: GTP, GDP |
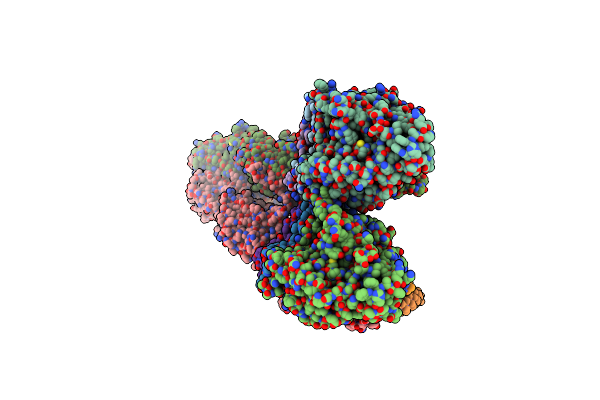 |
Structure Of The Y-Tubulin Small Complex (Ytusc) As Part Of The Native Y-Tubulin Ring Complex (Yturc) Capping Microtubule Minus Ends At The Spindle Pole Body
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-04-24 Classification: CELL CYCLE Ligands: GTP |
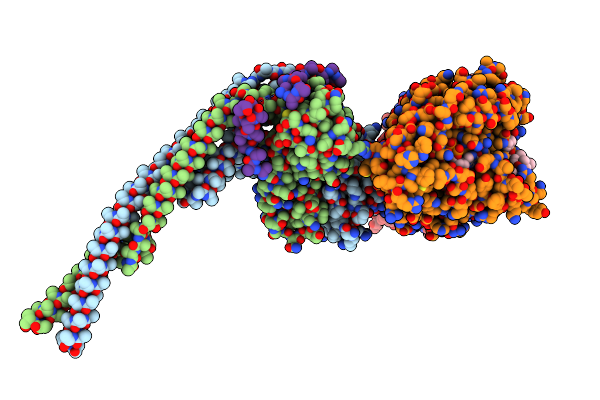 |
Organism: Saccharomyces cerevisiae, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2024-04-03 Classification: CELL CYCLE Ligands: GTP, MG, GDP, TA1 |
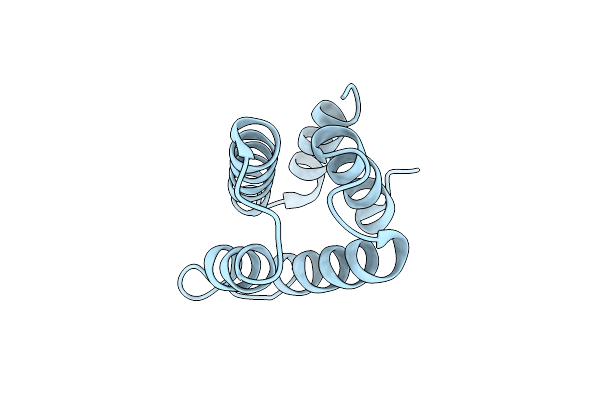 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2024-04-03 Classification: CELL CYCLE |
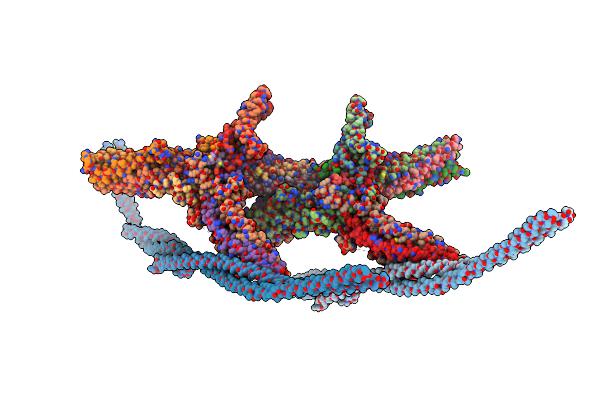 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-12-06 Classification: CELL CYCLE |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-12-06 Classification: CELL CYCLE |
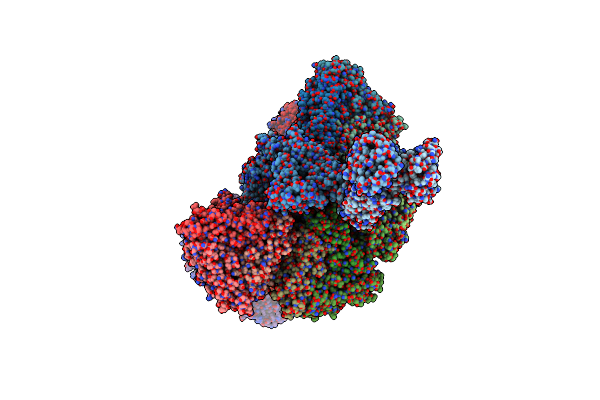 |
Organism: Rotavirus a
Method: ELECTRON MICROSCOPY Release Date: 2023-05-10 Classification: VIRAL PROTEIN |
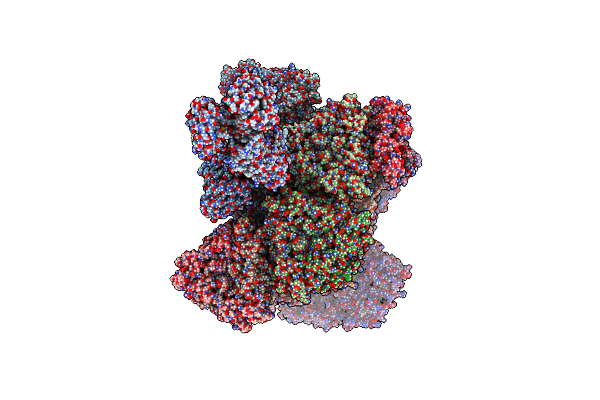 |
Organism: Rotavirus a
Method: ELECTRON MICROSCOPY Release Date: 2023-04-05 Classification: VIRAL PROTEIN Ligands: CA, ZN |
 |
Organism: Rotavirus a
Method: ELECTRON MICROSCOPY Release Date: 2023-04-05 Classification: VIRAL PROTEIN |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2021-03-03 Classification: STRUCTURAL PROTEIN Ligands: V8M |
 |
Native Structure Of Mosquitocidal Cyt1A Protoxin Obtained By Serial Femtosecond Crystallography On In Vivo Grown Crystals At Ph 7
Organism: Bacillus thuringiensis subsp. israelensis
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2020-10-14 Classification: TOXIN |
 |
Structure Of Mosquitocidal Cyt1A Protoxin Obtained By Serial Femtosecond Crystallography On In Vivo Grown Crystals Soaked With Dtt At Ph 7
Organism: Bacillus thuringiensis subsp. israelensis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-10-14 Classification: TOXIN |
 |
Structure Of Mosquitocidal Cyt1Aa Protoxin Obtained By Serial Femtosecond Crystallography On In Vivo Grown Crystals At Ph 10
Organism: Bacillus thuringiensis subsp. israelensis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-10-14 Classification: TOXIN Ligands: CA |
 |
Structure Of The C7S Mutant Of Mosquitocidal Cyt1A Protoxin Obtained By Serial Femtosecond Crystallography On In Vivo Grown Crystals At Ph 7
Organism: Bacillus thuringiensis subsp. israelensis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-10-14 Classification: TOXIN |
 |
Crystal Structure Of S. Aureus Penicillin Binding Protein 4 (Pbp4) Mutant (S75C)
Organism: Staphylococcus aureus (strain col)
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2018-12-19 Classification: HYDROLASE Ligands: ZN |

