Search Count: 8
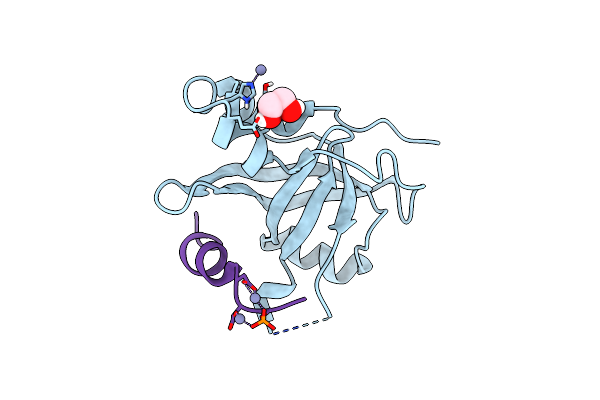 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2023-09-27 Classification: PROTEIN BINDING Ligands: EDO, ZN |
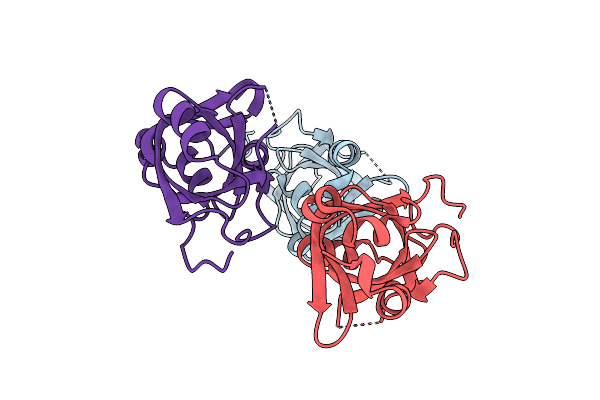 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-09-27 Classification: PROTEIN BINDING |
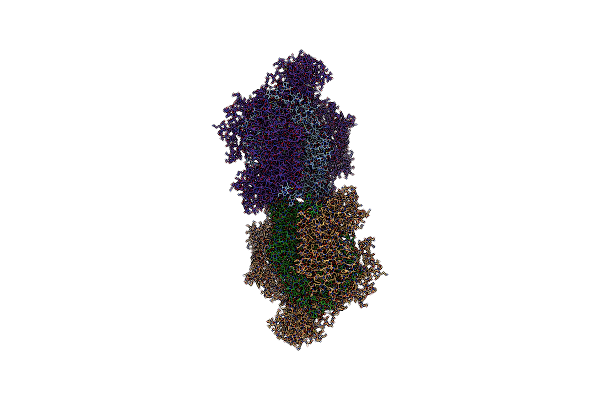 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2020-11-11 Classification: HYDROLASE Ligands: ANP, ZN, MG |
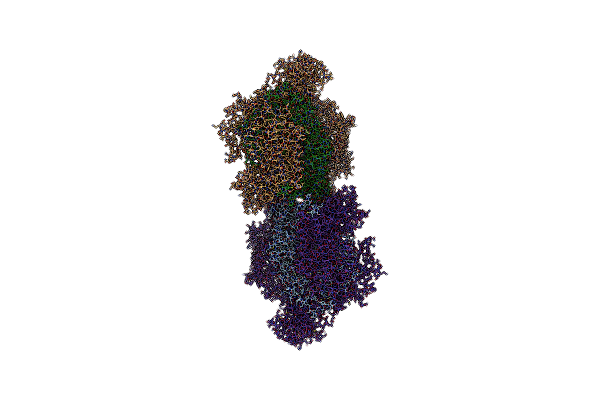 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2020-11-11 Classification: HYDROLASE Ligands: ANP, ZN, MG |
 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2020-11-11 Classification: HYDROLASE Ligands: ANP, ZN |
 |
T. Maritima Rnase H2 G21S In Complex With Nucleic Acid Substrate And Calcium Ions
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2013-02-06 Classification: Hydrolase/DNA Ligands: CA |
 |
Crystal Structure Of The Eukaryotic Dna Polymerase Processivity Factor Pcna
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 1995-03-31 Classification: DNA-BINDING Ligands: HG |
 |
Crystal Structure Of The Eukaryotic Dna Polymerase Processivity Factor Pcna
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 1995-03-31 Classification: DNA-BINDING |

