Search Count: 11
 |
Crystal Structure Of Domain-Of-Unknown-Function Duf4867 From Bacillus Megaterium
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-02-26 Classification: ISOMERASE Ligands: FE, NA, CL |
 |
Crystal Structure Of Domain-Of-Unknown-Function Duf4867 From Bacillus Megaterium (Unmodelled Additional Ligand Density At Active Site)
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-02-26 Classification: ISOMERASE Ligands: FE |
 |
Crystal Structure Of Sulfoquinovose-1-Dehydrogenase From Pseudomonas Putida (Sulfo-Ed Pathway)
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Sulfoquinovose-1-Dehydrogenase From Pseudomonas Putida In Complex With Nad+ (Sulfo-Ed Pathway)
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Crystal Structure Of Sulfoquinovose-1-Dehydrogenase From Pseudomonas Putida In Complex With Sulfoquinovose Substrate (Sulfo-Ed Pathway)
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: YZT |
 |
Organism: Cupriavidus pinatubonensis jmp134
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2024-09-25 Classification: OXIDOREDUCTASE Ligands: EDO, ZN |
 |
Crystal Structure Of Dhps-3-Dehydrogenase, Hpsn From Cupriavidus Pinatubonensis In Complex With Nadh
Organism: Cupriavidus pinatubonensis jmp134
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2024-09-25 Classification: OXIDOREDUCTASE Ligands: ZN, NAI |
 |
Crystal Structure Of Hpsn D352A Mutant From Cupriavidus Pinatubonensis In Complex With Nad+
Organism: Cupriavidus pinatubonensis jmp134
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2024-09-25 Classification: OXIDOREDUCTASE Ligands: NAD, ZN |
 |
Crystal Structure Of Dhps-3-Dehydrogenase, Hpsn From Cupriavidus Pinatubonensis In Complex With Product Analogue (L-Cysteate) And Nadh
Organism: Cupriavidus pinatubonensis jmp134
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-09-25 Classification: OXIDOREDUCTASE Ligands: ZN, NAI, OCS |
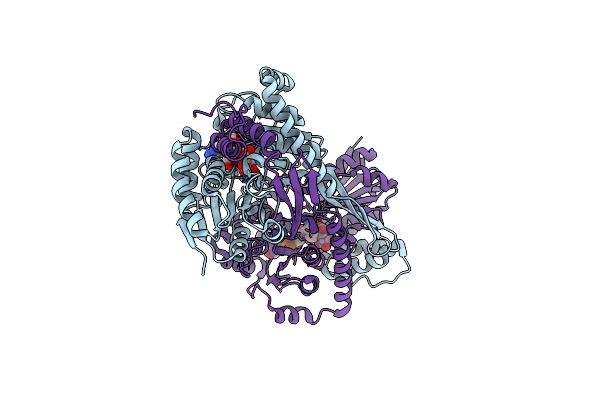 |
Crystal Structure Of Dhps-3-Dehydrogenase, Hpsn From Cupriavidus Pinatubonensis In Complex With Product (R-Sulfolactate) And Nadh
Organism: Cupriavidus pinatubonensis jmp134
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2024-09-25 Classification: OXIDOREDUCTASE Ligands: ZN, NAI, 3SL |
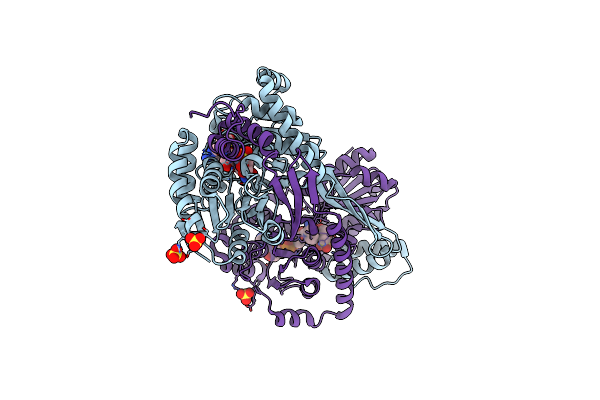 |
Crystal Structure Of Dhps-3-Dehydrogenase, Hpsn H319A Variant From Cupriavidus Pinatubonensis In Complex With Substrate (R-Dhps) And Nadh
Organism: Cupriavidus pinatubonensis jmp134
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2024-09-25 Classification: OXIDOREDUCTASE Ligands: ZN, NAI, A1AZH, SO4 |

