Search Count: 25
 |
Oxa-48_F72L. Epistasis Arises From Shifting The Rate-Limiting Step During Enzyme Evolution
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2024-02-14 Classification: HYDROLASE Ligands: CL |
 |
Oxa-48_Q5. Epistasis Arises From Shifting The Rate-Limiting Step During Enzyme Evolution
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.17 Å Release Date: 2024-02-14 Classification: HYDROLASE Ligands: CL, SO4, PEG, MLI |
 |
Oxa-48_Q5-Caz. Epistasis Arises From Shifting The Rate-Limiting Step During Enzyme Evolution
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2024-02-14 Classification: HYDROLASE Ligands: CTJ, CL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-08-09 Classification: ELECTRON TRANSPORT Ligands: HEM, LMR, CL |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2021-10-06 Classification: DE NOVO PROTEIN Ligands: HEM |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2021-04-28 Classification: DE NOVO PROTEIN Ligands: ZN, PEG, DMX, BEZ |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-04-21 Classification: DE NOVO PROTEIN Ligands: BEZ, 1PE, ZN, PEG, ACY |
 |
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2020-12-02 Classification: HYDROLASE |
 |
Crystal Structure Of Kemp Eliminase Hg3 In Complex With The Transition State Analog 6-Nitrobenzotriazole
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: 6NT, SO4 |
 |
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2020-12-02 Classification: HYDROLASE |
 |
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2020-12-02 Classification: HYDROLASE |
 |
Crystal Structure Of Kemp Eliminase Hg3 K50Q In Complex With The Transition State Analog 6-Nitrobenzotriazole
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: 6NT |
 |
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: PG4 |
 |
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Of Kemp Eliminase Hg3.7 In Complex With The Transition State Analog 6-Nitrobenzotriazole
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: 6NT, ACT, SO4 |
 |
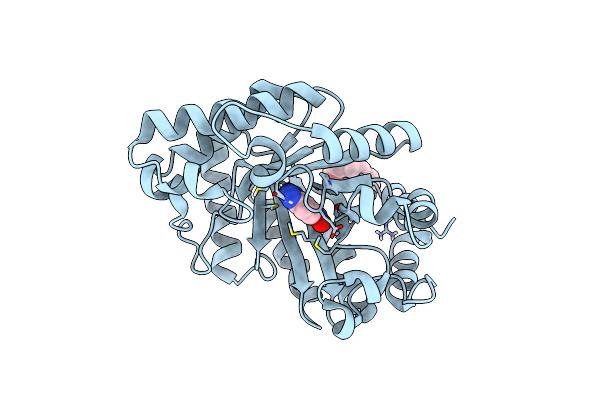
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-12-02 Classification: HYDROLASE |
 |
Crystal Structure Of Kemp Eliminase Hg3.17 In Complex With The Transition State Analog 6-Nitrobenzotriazole
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2020-12-02 Classification: HYDROLASE Ligands: 1PE, 6NT |
 |
Kemp Eliminase Hg3.17 Mutant Q50F, E47N, N300D Complexed With Transition State Analog 6-Nitrobenzotriazole
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2020-04-01 Classification: DE NOVO PROTEIN Ligands: GOL, 6NT |
 |
Kemp Eliminase Hg3.17 Mutant Q50S, E47N, N300D Complexed With Transition State Analog 6-Nitrobenzotriazole
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:0.95 Å Release Date: 2020-04-01 Classification: DE NOVO PROTEIN Ligands: 6NT, SO4 |

