Search Count: 18
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:0.81 Å Release Date: 2016-03-09 Classification: STRUCTURAL PROTEIN Ligands: DMS |
 |
Organism: Lactobacillus salivarius
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-10-08 Classification: TRANSFERASE |
 |
Thiamine Pyrophosphate Bound Transketolase From Lactobacillus Salivarius At 2.2A Resolution
Organism: Lactobacillus salivarius
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2014-10-08 Classification: TRANSFERASE Ligands: MG, TPP |
 |
Crystal Structure Of A Hd-Gyp Domain (A Cyclic-Di-Gmp Phosphodiesterase) Containing A Tri-Nuclear Metal Centre
Organism: Persephonella marina
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2014-02-19 Classification: HYDROLASE Ligands: FE, SIN, IMD, EDO |
 |
Crystal Structure Of A Hd-Gyp Domain (A Cyclic-Di-Gmp Phosphodiesterase) Containing A Tri-Nuclear Metal Centre
Organism: Persephonella marina
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2014-02-19 Classification: HYDROLASE Ligands: FE, SIN, C2E |
 |
Crystal Structure Of A Hd-Gyp Domain (A Cyclic-Di-Gmp Phosphodiesterase) Containing A Tri-Nuclear Metal Centre
Organism: Persephonella marina
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2014-02-19 Classification: HYDROLASE Ligands: FE, SIN, IMD, 5GP |
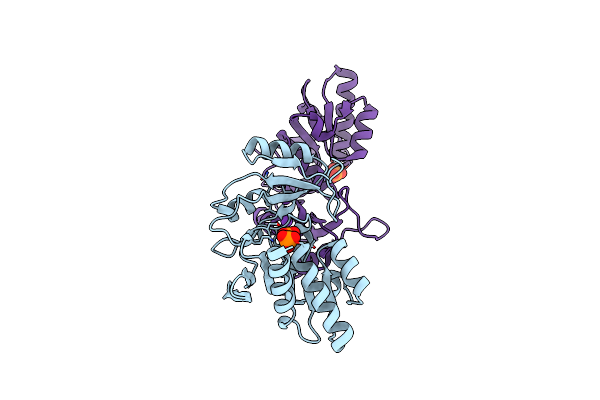 |
Crystal Structure Of Ribose 5-Phosphate Isomerase From The Probiotic Bacterium Lactobacillus Salivarius Ucc118
Organism: Lactobacillus salivarius
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2012-08-29 Classification: ISOMERASE Ligands: PO4, K |
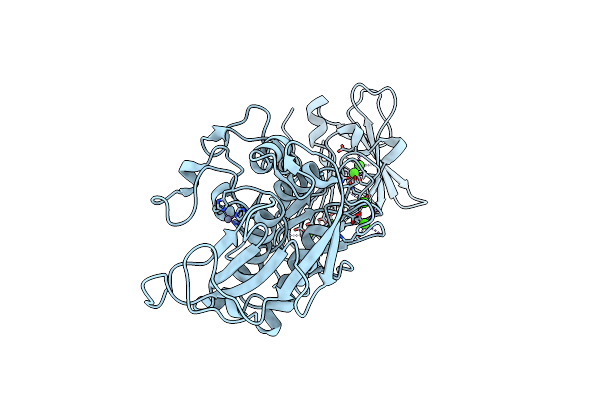 |
Organism: Erwinia chrysanthemi
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2009-08-04 Classification: HYDROLASE Ligands: CA, ZN, CL |
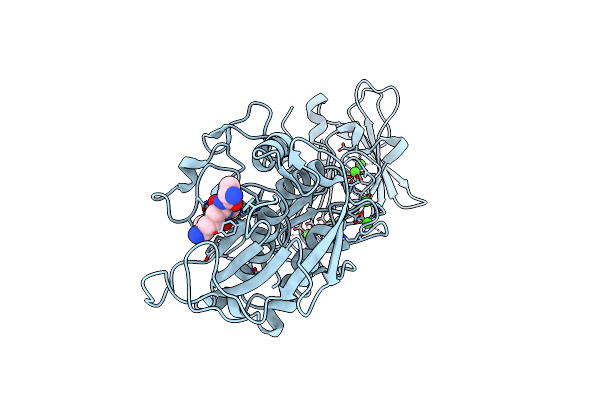 |
Organism: Erwinia chrysanthemi, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2009-07-21 Classification: HYDROLASE Ligands: CA, ZN, CL |
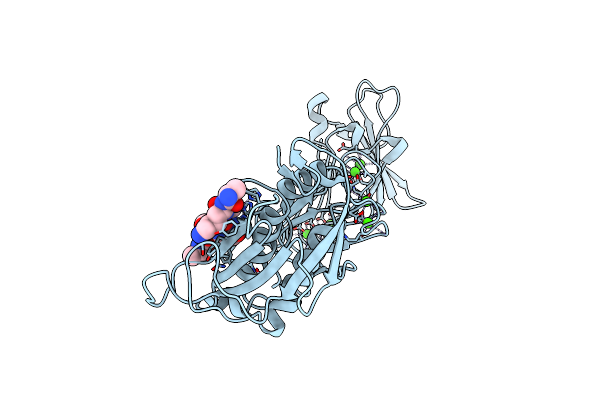 |
Organism: Erwinia chrysanthemi, Unidentified
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2009-06-30 Classification: HYDROLASE Ligands: CA, ZN, CL |
 |
Organism: Erwinia chrysanthemi, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2009-06-30 Classification: HYDROLASE Ligands: CA, CL |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2008-06-24 Classification: HYDROLASE/TRANSLATION Ligands: AMP |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2008-06-24 Classification: TRANSLATION/HYDROLASE Ligands: AMP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2006-06-01 Classification: TRANSCRIPTION |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2006-02-09 Classification: CELL DIVISION PROTEIN Ligands: ZN, ADP, MG |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2006-02-09 Classification: HYDROLASE Ligands: ZN, ADP, MG |
 |
Crystal Structure Of The Phosphoenolpyruvate-Binding Enzyme I-Domain From The Thermoanaerobacter Tengcongensis Pep: Sugar Phosphotransferase System (Pts)
Organism: Thermoanaerobacter tengcongensis
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2005-02-02 Classification: TRANSFERASE |
 |
Crystal Structure Of Yeast Ypr118W, A Methylthioribose-1-Phosphate Isomerase Related To Regulatory Eif2B Subunits
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2004-07-16 Classification: ISOMERASE Ligands: SO4 |

