Search Count: 42
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-07-10 Classification: TRANSFERASE Ligands: ADP |
 |
Structure Of The Rat Vesicular Glutamate Transporter 2 Determined By Single-Particle Cryo-Em
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: MEMBRANE PROTEIN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-05-26 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2021-05-26 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
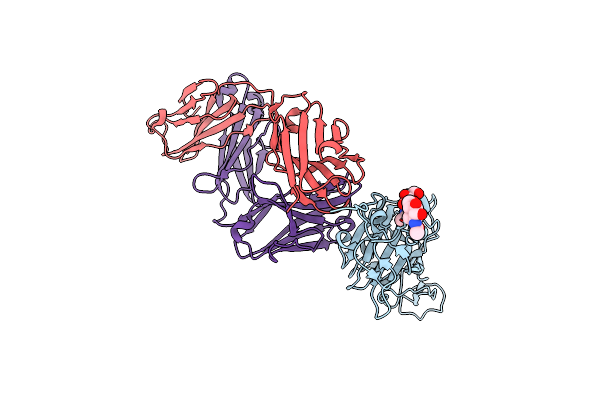
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-05-26 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-05-26 Classification: VIRAL PROTEIN Ligands: NAG |
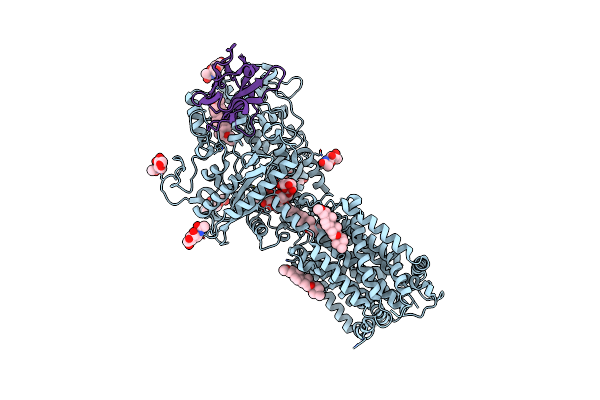 |
Hedgehog Receptor Patched (Ptch1) In Complex With Conformation Selective Nanobody Ti23
Organism: Mus musculus, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2021-03-17 Classification: MEMBRANE PROTEIN Ligands: NAG, Q7G |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2020-12-16 Classification: OXIDOREDUCTASE |
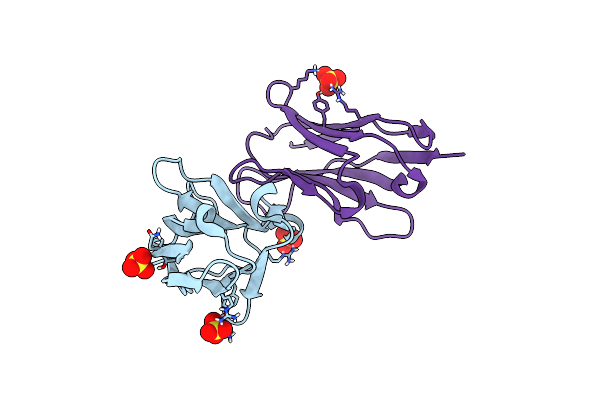 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2020-11-25 Classification: IMMUNE SYSTEM Ligands: SO4, CL |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2020-11-11 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2020-11-11 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Structural Basis For Cholesterol Transport-Like Activity Of The Hedgehog Receptor Patched
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2018-11-28 Classification: MEMBRANE PROTEIN Ligands: CLR |
 |
Cryo-Em Structure Of The Tmem16A Calcium-Activated Chloride Channel In Nanodisc
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2017-12-27 Classification: MEMBRANE PROTEIN Ligands: CA |
 |
Cryo-Em Structure Of The Tmem16A Calcium-Activated Chloride Channel In Lmng
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2017-12-27 Classification: MEMBRANE PROTEIN Ligands: CA |
 |
Structure Of A Mechanotransduction Ion Channel Drosophila Nompc In Nanodisc
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2017-06-28 Classification: MEMBRANE PROTEIN Ligands: PCF |
 |
Cryo-Em Structure Of Polycystic Kidney Disease Protein 2 (Pkd2), Residues 198-703
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2016-11-02 Classification: METAL TRANSPORT Ligands: NAG |
 |
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Elongation Factor G In The Pre-Translocational State
Organism: Escherichia coli str. k-12 substr. mc4100, Thermus thermophilus, Synthetic construct, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2015-01-28 Classification: RIBOSOME Ligands: MG, K, ZN, SF4, GDP |
 |
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Elongation Factor G And Fusidic Acid In The Post-Translocational State
Organism: Escherichia coli str. k-12 substr. mc4100, Thermus thermophilus, Synthetic construct, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2015-01-28 Classification: RIBOSOME Ligands: MG, ZN, SF4, FUA, GDP |
 |
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Elongation Factor G Trapped By The Antibiotic Dityromycin
Organism: Escherichia coli str. k-12 substr. mc4100, Thermus thermophilus, Synthetic construct, Streptomyces sp., Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2015-01-28 Classification: Ribosome/antibiotic Ligands: MG, ZN, SF4, GDP |
 |
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Elongation Factor G In The Post-Translocational State (Without Fusitic Acid)
Organism: Escherichia coli str. k-12 substr. mc4100, Thermus thermophilus, Synthetic construct, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2015-01-28 Classification: RIBOSOME Ligands: MG, K, ZN, SF4, GDP |

