Search Count: 69
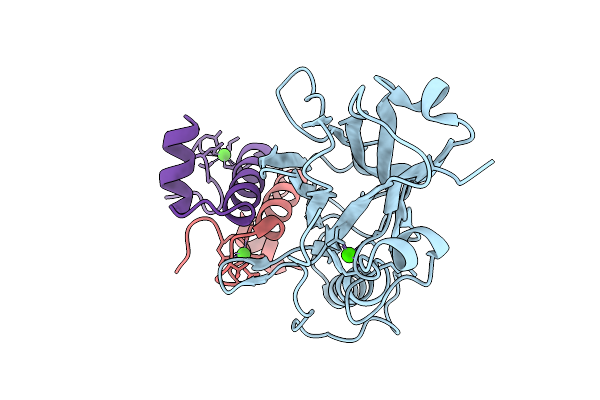 |
Ruminococcus Flavefaciens Coh-Doc Complex Between A Group 2 Dockerin And The Cohesin From Cell Surface Attached Scaffoldin Scae
Organism: Ruminococcus flavefaciens fd-1
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: PROTEIN BINDING Ligands: CA |
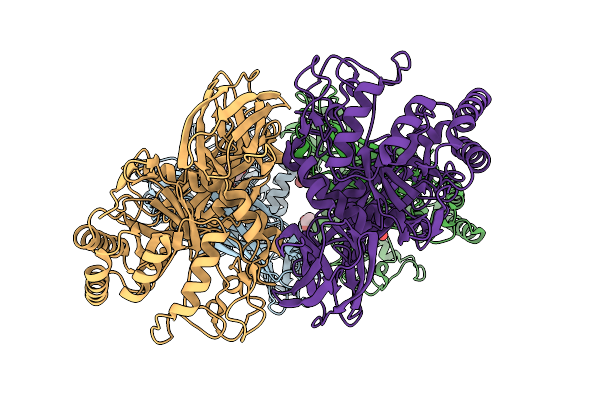 |
Glycoside Hydrolase Family 157 From Labilibaculum Antarcticum, Wild Type Semet Derivative (Lagh157)
Organism: Labilibaculum antarcticum
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: HYDROLASE Ligands: ACT, GOL, MLA |
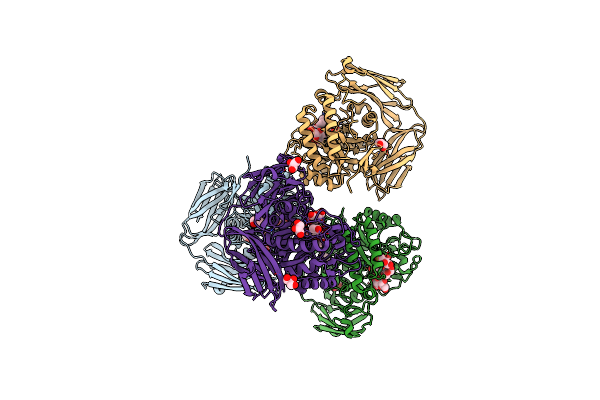 |
Glycoside Hydrolase Family 157 From Labilibaculum Antarcticum (Lagh157) E224A Mutant In Complex With Laminaritriose And Glucose
Organism: Labilibaculum antarcticum
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2025-05-21 Classification: HYDROLASE Ligands: EDO, BGC, GOL, PGE |
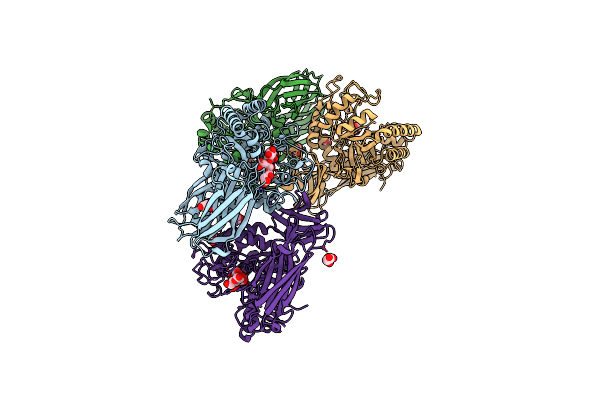 |
Glycoside Hydrolase Family 157 From Labilibaculum Antarcticum (Lagh157) In Complex With Laminaribiose
Organism: Labilibaculum antarcticum
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2025-05-21 Classification: HYDROLASE Ligands: MLA, GOL, BGC, PG4 |
 |
Crystal Structure Of The Dimeric C-Terminal Big_2-Cbm56 Domains From Paenibacillus Illinoisensis (Bacillus Circulans Iam1165) Beta-1,3-Glucanase H
Organism: Paenibacillus illinoisensis
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2023-02-22 Classification: HYDROLASE Ligands: PG4, GOL, PG6, CL |
 |
Crystal Structure Of The Tetrameric C-Terminal Big_2-Cbm56 Domains From Paenibacillus Illinoisensis (Bacillus Circulans Iam1165) Beta-1,3-Glucanase H
Organism: Paenibacillus illinoisensis
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2023-02-15 Classification: HYDROLASE Ligands: PG4, PEG, GOL, CL, MG |
 |
Crystal Structure Of The Semet Octameric C-Terminal Big_2-Cbm56 Domains From Paenibacillus Illinoisensis (Bacillus Circulans Iam1165) Beta-1,3-Glucanase H
Organism: Paenibacillus illinoisensis
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2023-02-01 Classification: HYDROLASE Ligands: GOL, CL |
 |
Ruminococcus Flavefaciens Cohesin-Dockerin Structure: Dockerin From Scah Adaptor Scaffoldin In Complex With The Cohesin From Scae Anchoring Scaffoldin
Organism: Ruminococcus flavefaciens fd-1
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2022-11-02 Classification: PROTEIN BINDING Ligands: CA, GOL, SO4 |
 |
Structure Of D165A/D167A Double Mutant Of Chit33 From Trichoderma Harzianum Complexed With Chitintetraose.
Organism: Trichoderma harzianum
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-08-31 Classification: HYDROLASE |
 |
Organism: Trichoderma harzianum
Method: X-RAY DIFFRACTION Resolution:1.12 Å Release Date: 2022-08-31 Classification: HYDROLASE Ligands: TRS, PEG, GOL |
 |
Crystal Structure Of E. Coli Beta-Glucuronidase In Complex With Covalent Inhibitor Me727
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2022-08-03 Classification: CARBOHYDRATE Ligands: 8I4 |
 |
Crystal Structure Of Human Heparanase In Complex With Covalent Inhibitor Vl166
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2022-08-03 Classification: CARBOHYDRATE Ligands: NAG, EDO, CL |
 |
Crystal Structure Of Human Heparanase In Complex With Covalent Inhibitor Gr109
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2022-08-03 Classification: CARBOHYDRATE Ligands: NAG, EDO, CL |
 |
Crystal Structure Of Burkholderia Pseudomallei Heparanase In Complex With Covalent Inhibitor Vl166
Organism: Burkholderia pseudomallei (strain k96243)
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2022-08-03 Classification: HYDROLASE Ligands: EDO |
 |
Crystal Structure Of Burkholderia Pseudomallei Heparanase In Complex With Covalent Inhibitor Gr109
Organism: Burkholderia pseudomallei (strain k96243)
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2022-08-03 Classification: HYDROLASE Ligands: EDO |
 |
Crystal Structure Of Human Heparanase In Complex With Covalent Inhibitor Cb678
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-08-03 Classification: CARBOHYDRATE Ligands: NAG, EDO, CL |
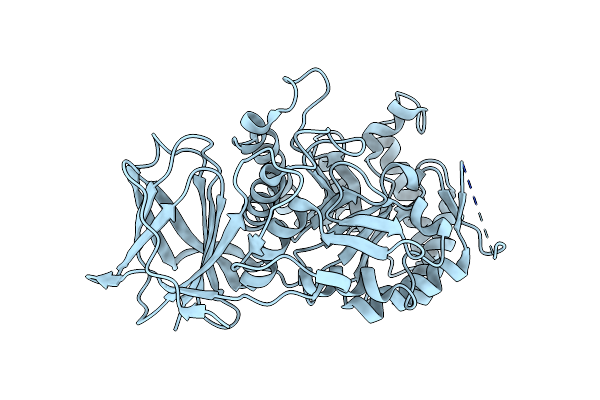 |
Crystal Structure Of Beta-Glucuronidase From Acidobacterium Capsulatum At 1.24 Angstrom Resolution
Organism: Acidobacterium capsulatum (strain atcc 51196 / dsm 11244 / jcm 7670 / nbrc 15755 / ncimb 13165 / 161)
Method: X-RAY DIFFRACTION Resolution:1.24 Å Release Date: 2022-08-03 Classification: CARBOHYDRATE |
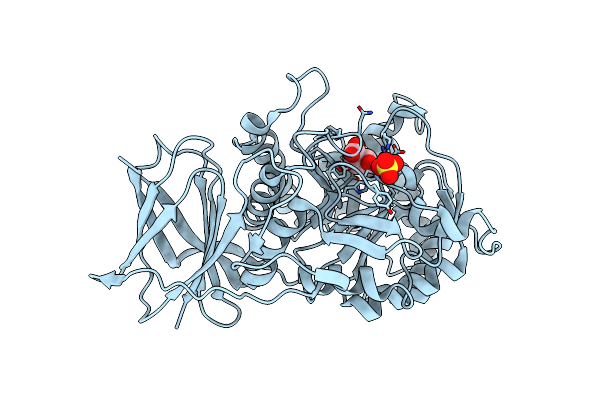 |
Crystal Structure Of Beta-Glucuronidase From Acidobacterium Capsulatum In Complex With Covalent Inhibitor Me727
Organism: Acidobacterium capsulatum (strain atcc 51196 / dsm 11244 / jcm 7670 / nbrc 15755 / ncimb 13165 / 161)
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2022-08-03 Classification: CARBOHYDRATE Ligands: 8I4, SO4 |
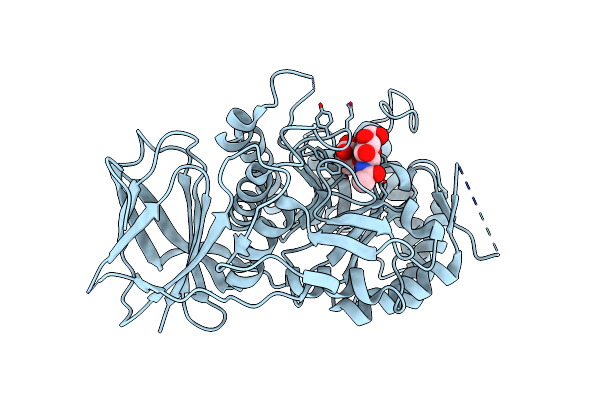 |
Crystal Structure Of Beta-Glucuronidase From Acidobacterium Capsulatum In Complex With Covalent Inhibitor Vl166
Organism: Acidobacterium capsulatum (strain atcc 51196 / dsm 11244 / jcm 7670 / nbrc 15755 / ncimb 13165 / 161)
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2022-08-03 Classification: CARBOHYDRATE |
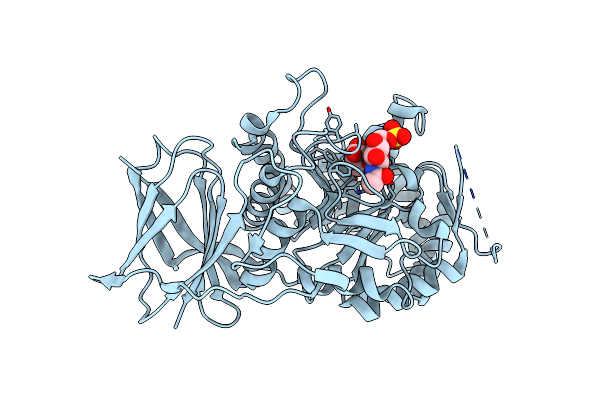 |
Crystal Structure Of Beta-Glucuronidase From Acidobacterium Capsulatum In Complex With Covalent Inhibitor Gr109
Organism: Acidobacterium capsulatum (strain atcc 51196 / dsm 11244 / jcm 7670 / nbrc 15755 / ncimb 13165 / 161)
Method: X-RAY DIFFRACTION Resolution:1.09 Å Release Date: 2022-08-03 Classification: CARBOHYDRATE |

