Search Count: 70
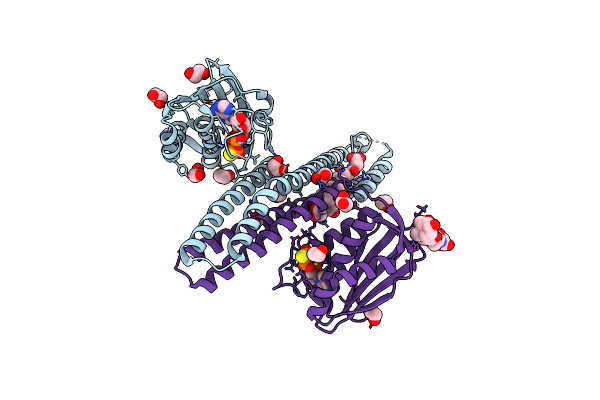 |
Crystal Structure Of Hprs Histidine Kinase Cytoplasmic Fragment From Escherichia Coli
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-01-22 Classification: TRANSFERASE Ligands: AGS, MG, GCH, EDO, IMD |
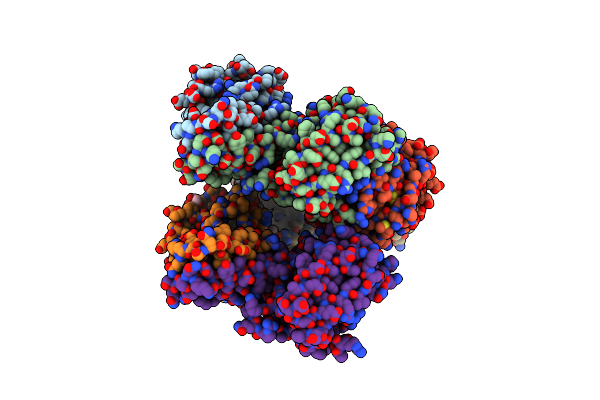 |
Crystal Structure Of Cuss Histidine Kinase Catalytic Core From Escherichia Coli
Organism: Escherichia coli 'bl21-gold(de3)plyss ag'
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-08-31 Classification: TRANSFERASE Ligands: EDO |
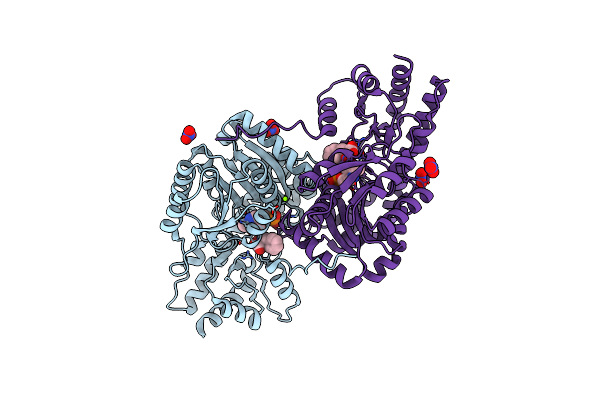 |
Psychrophilic Aromatic Amino Acids Aminotransferase From Psychrobacter Sp. B6 Cocrystalized With Substrate Analog - L-(-)-3-Phenyllactic Acid
Organism: Psychrobacter sp. b6
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-07-14 Classification: TRANSFERASE Ligands: MG, PLP, NO3, HFA |
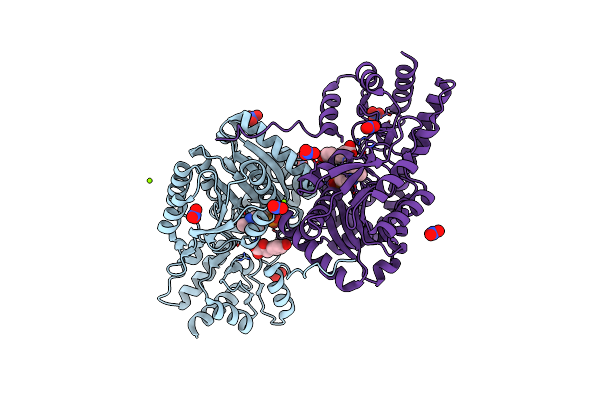 |
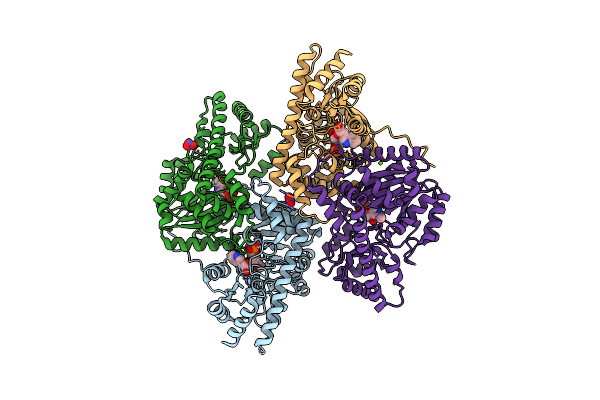
Psychrophilic Aromatic Amino Acids Aminotransferase From Psychrobacter Sp. B6 Cocrystalized With Substrate Analog - L-P-Hydroxyphenyllactic Acid
Organism: Psychrobacter sp. b6
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2021-07-14 Classification: TRANSFERASE Ligands: MG, TYF, NO3, PLP |
 |
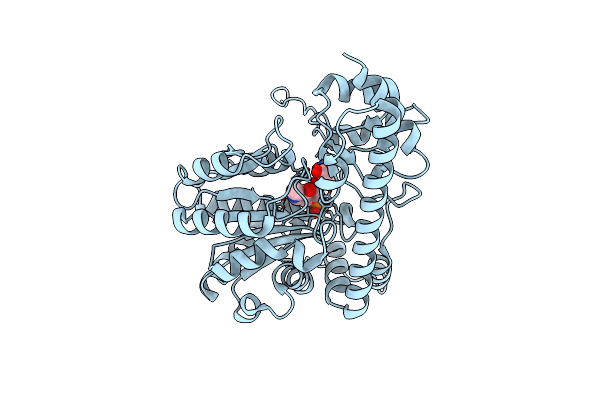
Psychrophilic Aromatic Amino Acids Aminotransferase From Psychrobacter Sp. B6 Cocrystalized With Substrate Analog - L-Indole-3-Lactic Acid
Organism: Psychrobacter sp. b6
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2021-07-14 Classification: TRANSFERASE Ligands: MG, NO3, PLP, 3IL |
 |
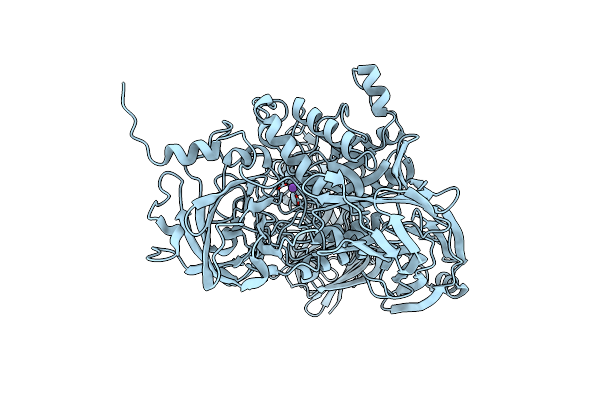
Psychrophilic Aromatic Amino Acids Aminotransferase From Psychrobacter Sp. B6 Cocrystalized With Substrate Analog - Malic Acid
Organism: Psychrobacter sp. b6
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2020-11-18 Classification: TRANSFERASE Ligands: PLP, LMR |
 |
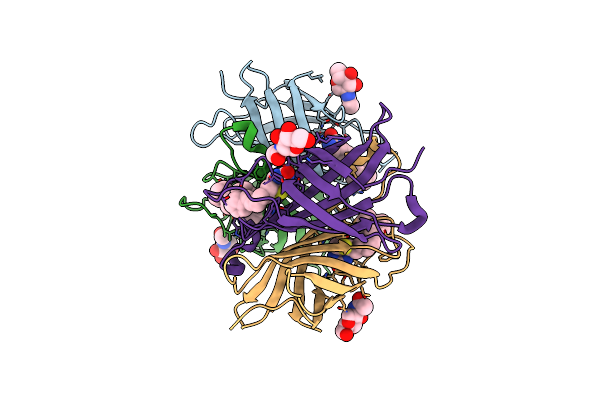
Cold-Adapted Beta-D-Galactosidase From Arthrobacter Sp. 32Cb - Data Collected At Room Temperature
Organism: Arthrobacter sp. 32cb
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2019-06-26 Classification: HYDROLASE |
 |
Crystal Structure Of Avidin In Complex With Ferrocene Homobiotin Derivative
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2018-02-14 Classification: Biotin-binding protein Ligands: HBF, NAG, IOD |
 |
Organism: Trametes sanguinea
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2017-11-01 Classification: OXIDOREDUCTASE Ligands: CU, PER |
 |
Organism: Pycnoporus coccineus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-11-01 Classification: OXIDOREDUCTASE Ligands: CU, PER, ME2, IPH, NA, MLI |
 |
Crystal Structure Of Laccases From Pycnoporus Sanguineus, Izoform Ii, Monoclinic
Organism: Trametes sanguinea
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2017-11-01 Classification: OXIDOREDUCTASE Ligands: CU, PER, PEG, FMT, NAG |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-01-18 Classification: BIOTIN-BINDING PROTEIN Ligands: B9F, NAG |
 |
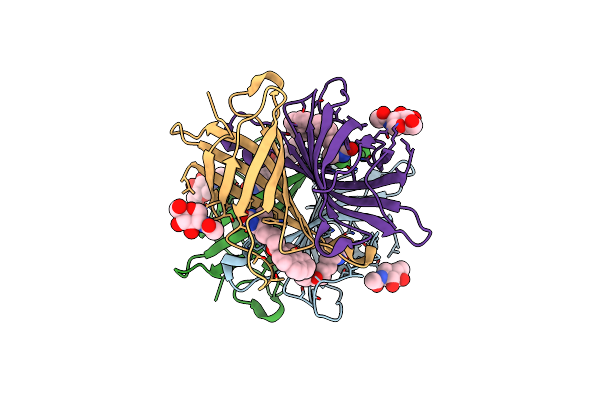
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-10-12 Classification: biotin binding protein Ligands: NAG, B9P |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-10-12 Classification: biotin binding protein Ligands: NAG, D9P |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-03-23 Classification: GLYCOPROTEIN Ligands: NAG, 55R |
 |
Organism: Candida antarctica
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-01-13 Classification: HYDROLASE |
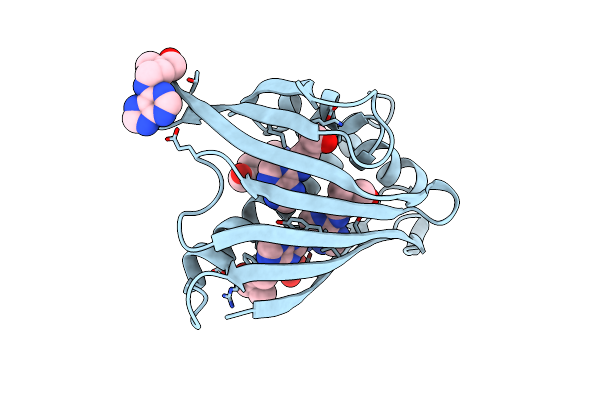 |
Crystal Structure Of Yellow Lupin Llpr-10.1A Protein In Complex With Trans-Zeatin
Organism: Lupinus luteus
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2015-12-09 Classification: PLANT PROTEIN Ligands: ZEA, SO4 |
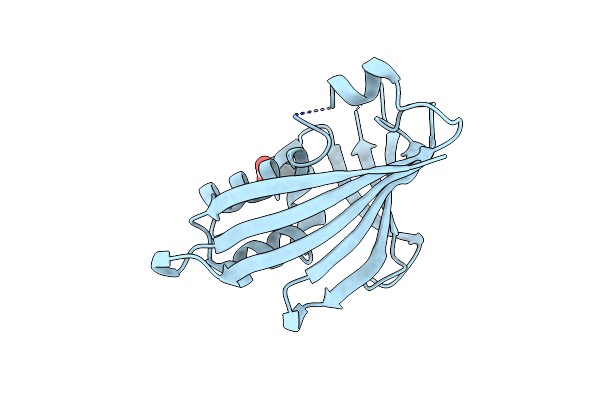 |
Organism: Lupinus luteus
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2015-12-09 Classification: PLANT PROTEIN Ligands: ACT |
 |
Crystal Structure Of Yellow Lupine Llpr-10.1A Protein Partially Saturated With Trans-Zeatin
Organism: Lupinus luteus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2015-12-09 Classification: PLANT PROTEIN |
 |
Organism: Bombyx mori
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-03-04 Classification: LIPID BINDING PROTEIN Ligands: MES, EDO, PEG, IPA |

