Search Count: 35
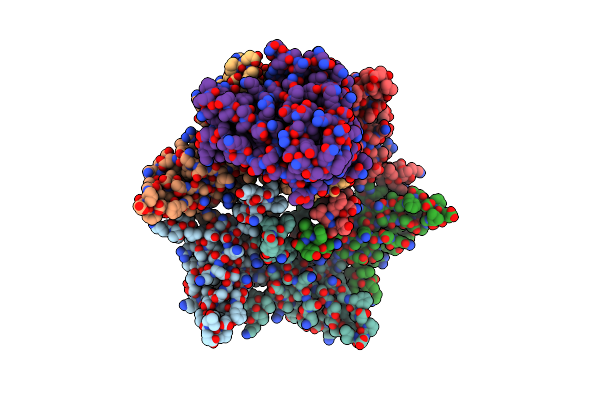 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-07-10 Classification: LIPID BINDING PROTEIN |
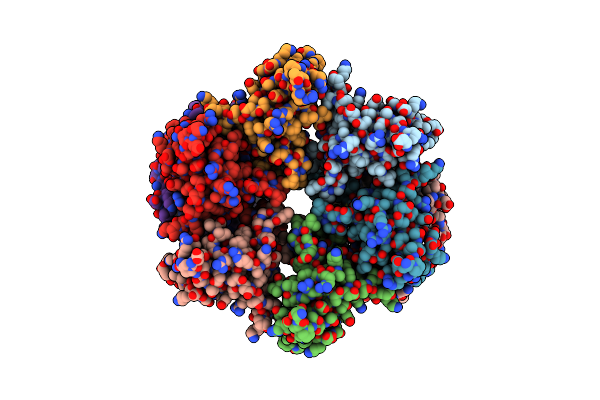 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-07-10 Classification: LIPID TRANSPORT |
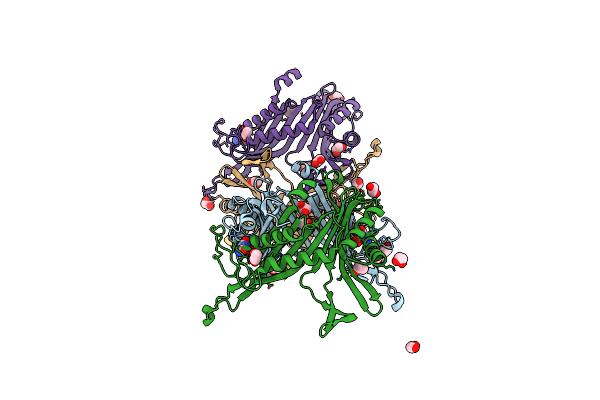 |
Crystal Structure Of The Cysteine-Rich Gallus Gallus Urate Oxidase In Complex With The 8-Azaxanthine Inhibitor Under Reducing Conditions (Space Group C 2 2 21)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2024-01-17 Classification: OXIDOREDUCTASE Ligands: AZA, OXY, EDO, CL |
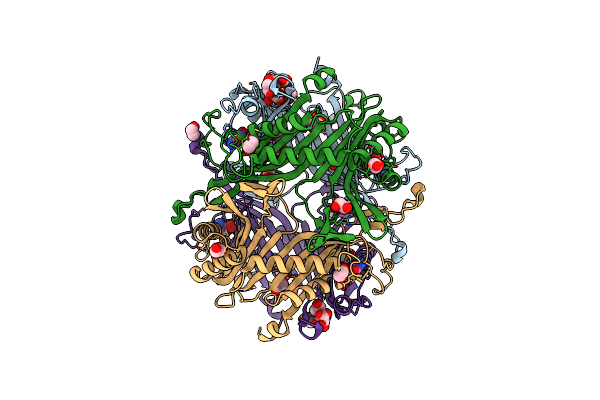 |
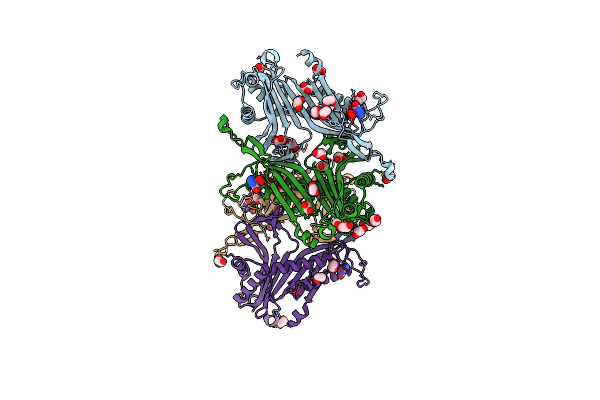
Crystal Structure Of The Cysteine-Rich Gallus Gallus Urate Oxidase In Complex With The 8-Azaxanthine Inhibitor Under Reducing Conditions (Space Group P 21 21 21)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2024-01-17 Classification: OXIDOREDUCTASE Ligands: AZA, EDO, TAR, OXY, CL |
 |
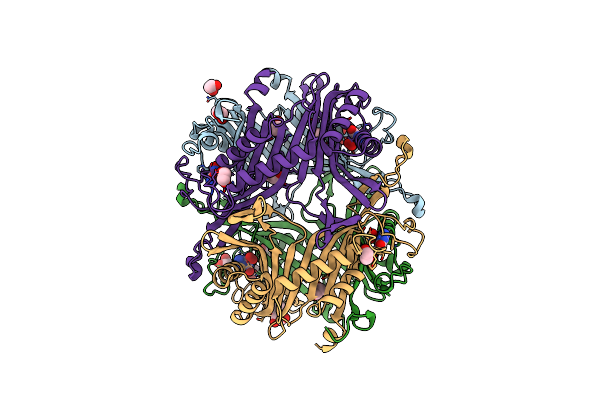
Crystal Structure Of The Cysteine-Rich Gallus Gallus Urate Oxidase In Complex With The 8-Azaxanthine Inhibitor Under Oxidising Conditions (Space Group C 2 2 21)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2024-01-17 Classification: OXIDOREDUCTASE Ligands: AZA, OXY, EDO, CL, BR |
 |
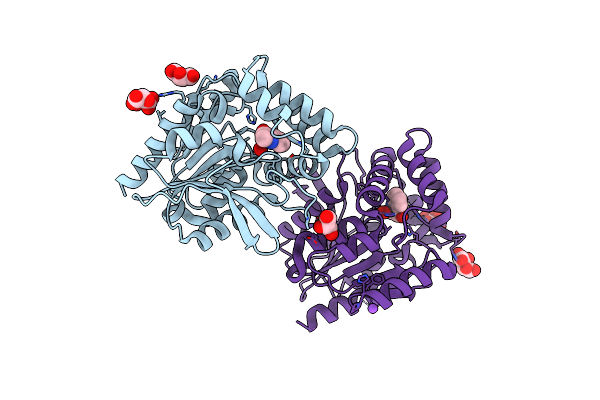
Crystal Structure Of The Cysteine-Rich Gallus Gallus Urate Oxidase In Complex With The 8-Azaxanthine Inhibitor Under Oxidising Conditions (Space Group P 21 21 21)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2024-01-17 Classification: OXIDOREDUCTASE Ligands: AZA, OXY, EDO, CL |
 |
Crystal Structure Of The Cofactor-Devoid 1-H-3-Hydroxy-4- Oxoquinaldine 2,4-Dioxygenase (Hod) S101A Variant Complexed With 2-Methyl-Quinolin-4(1H)-One Under Hyperoxyc Conditions
Organism: Paenarthrobacter nitroguajacolicus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-01-17 Classification: OXIDOREDUCTASE Ligands: VFH, OXY, TAR, GOL, NA |
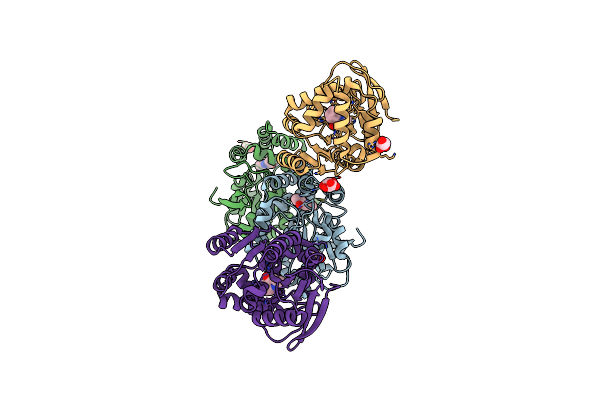 |
Crystal Structure Of The Cofactor-Devoid 1-H-3-Hydroxy-4- Oxoquinaldine 2,4-Dioxygenase (Hod) S101A Variant Complexed With 2-Methyl-Quinolin-4(1H)-One Under Normoxyc Conditions
Organism: Paenarthrobacter nitroguajacolicus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-01-17 Classification: OXIDOREDUCTASE Ligands: VFH, GOL, SRT |
 |
Crystal Structure Of The Cofactor-Devoid 1-H-3-Hydroxy-4- Oxoquinaldine 2,4-Dioxygenase (Hod) H251A Variant Complexed With N-Acetylanthranilate As Result Of In Crystallo Turnover Of Its Natural Substrate 1-H-3-Hydroxy-4- Oxoquinaldine Under Hyperoxic Conditions
Organism: Paenarthrobacter nitroguajacolicus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-01-17 Classification: OXIDOREDUCTASE Ligands: ZZ8, SRT, GOL, K |
 |
Room Temperature Crystal Structure Of The Cofactor-Devoid 1-H-3-Hydroxy-4- Oxoquinaldine 2,4-Dioxygenase (Hod) Under Xenon Pressure (30 Bar)
Organism: Paenarthrobacter nitroguajacolicus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2023-07-05 Classification: OXIDOREDUCTASE Ligands: XE, TAR |
 |
Serial Crystallography Structure Of Cofactor-Free Urate Oxidase In Complex With The 5-Peroxo Derivative Of 9-Methyl Uric Acid At Room Temperature
Organism: Aspergillus flavus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-04-26 Classification: OXIDOREDUCTASE Ligands: XDS |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-11-16 Classification: HYDROLASE Ligands: SO4 |
 |
Room Temperature Ssx Structure Of Gh11 Xylanase From Nectria Haematococca (1000 Frames)
Organism: Fusarium haematococcum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-11-16 Classification: HYDROLASE |
 |
Room Temperature Ssx Structure Of Gh11 Xylanase From Nectria Haematococca (40000 Frames)
Organism: Fusarium haematococcum
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2022-11-16 Classification: HYDROLASE |
 |
Room Temperature Ssx Structure Of Gh11 Xylanase From Nectria Haematococca (10000 Frames)
Organism: Fusarium haematococcum
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2022-11-16 Classification: HYDROLASE |
 |
Room Temperature Ssx Structure Of Gh11 Xylanase From Nectria Haematococca (4000 Frames)
Organism: Fusarium haematococcum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-11-16 Classification: HYDROLASE |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2022-11-16 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2022-11-16 Classification: HYDROLASE Ligands: SO4 |
 |
Crystal Structure Of The Cofactor-Devoid 1-H-3-Hydroxy-4- Oxoquinaldine 2,4-Dioxygenase (Hod) Catalytically Inactive H251A Variant Complexed With 2-Methyl-Quinolin-4(1H)-One Under Normoxic Conditions
Organism: Paenarthrobacter nitroguajacolicus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-06-01 Classification: OXIDOREDUCTASE Ligands: VFH, K, GOL, SRT, NA |
 |
Crystal Structure Of The Cofactor-Devoid 1-H-3-Hydroxy-4- Oxoquinaldine 2,4-Dioxygenase (Hod) Catalytically Inactive H251A Variant Complexed With 2-Methyl- Quinolin-4(1H)-One Under Hyperoxic Conditions
Organism: Paenarthrobacter nitroguajacolicus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-06-01 Classification: OXIDOREDUCTASE Ligands: VFH, K, TAR, GOL |

