Search Count: 9
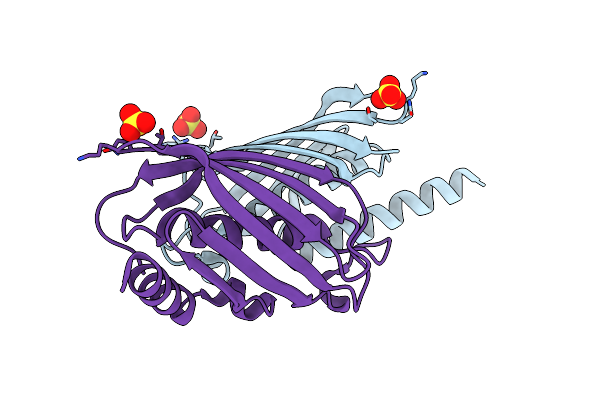 |
Structure Of The E. Coli Paai Protein From The Phyenylacetic Acid Degradation Operon
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-02-07 Classification: HYDROLASE Ligands: SO4 |
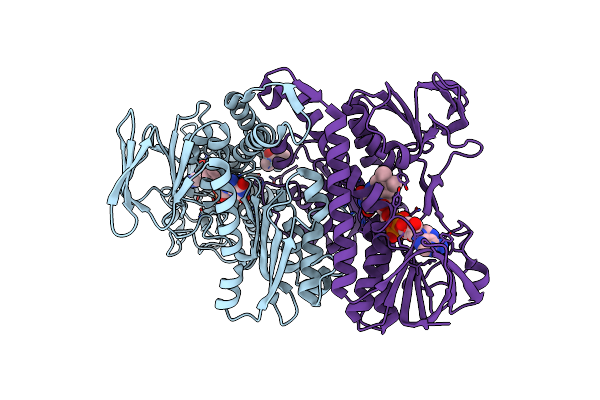 |
Crystal Structure Of Lipoamide Dehydrogenase From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2005-08-16 Classification: OXIDOREDUCTASE Ligands: FAD, MPD |
 |
Structure Of The E. Coli Paai Protein From The Phyenylacetic Acid Degradation Operon
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2003-07-08 Classification: HYDROLASE |
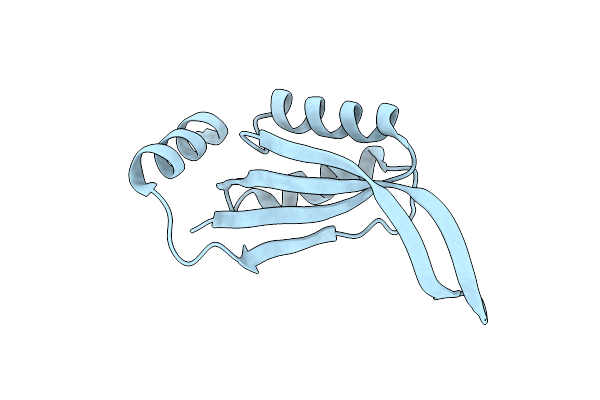 |
Structure Of The Periplasmic Divalent Cation Tolerance Protein Cuta From Archaeoglobus Fulgidus
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2003-04-29 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
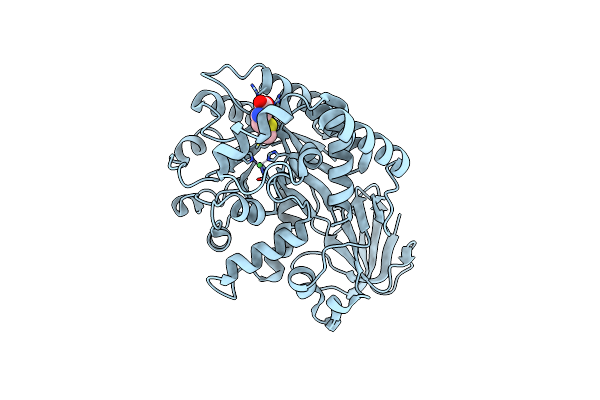 |
Structure Of Thermotoga Maritima Amidohydrolase Tm0936 Bound To Ni And Methionine
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2003-04-29 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION Ligands: NI, MET |
 |
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2003-01-07 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2002-06-12 Classification: TRANSFERASE |
 |
Structural Analysis Of Saccharomyces Cerevisiae Myo-Inositol Phosphate Synthase
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2002-04-10 Classification: ISOMERASE Ligands: NAD |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2001-08-22 Classification: OXIDOREDUCTASE, LYASE Ligands: FE2 |

