Search Count: 18
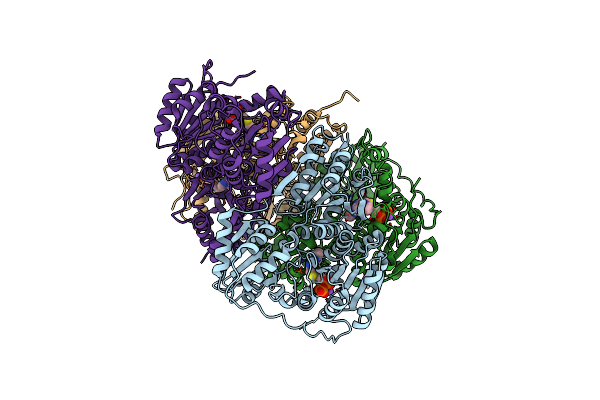 |
Rhodococcus Jostii Rha1 Thiamine Diphosphate-Dependent 4-Hydroxybenzoylformate Decarboxylase
Organism: Rhodococcus jostii (strain rha1)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2021-06-16 Classification: OXIDOREDUCTASE Ligands: TPP, NA |
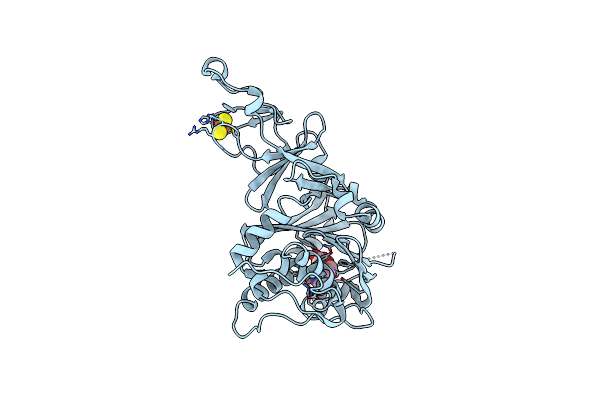 |
The Structure Of A Quaternary Ammonium Rieske Monooxygenase Reveals Insights Into Carnitine Oxidation By Gut Microbiota And Inter-Subunit Electron Transfer
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-03-31 Classification: OXIDOREDUCTASE Ligands: FE, 152, SCN, FES |
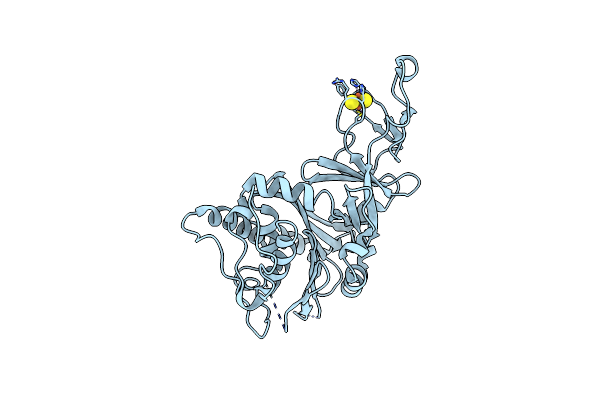 |
Crystal Structure Of The Apo Form Of A Quaternary Ammonium Rieske Monooxygenase Cnta
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2020-11-18 Classification: OXIDOREDUCTASE Ligands: FES |
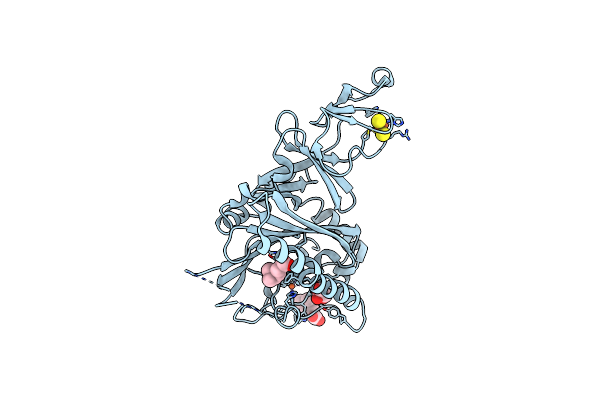 |
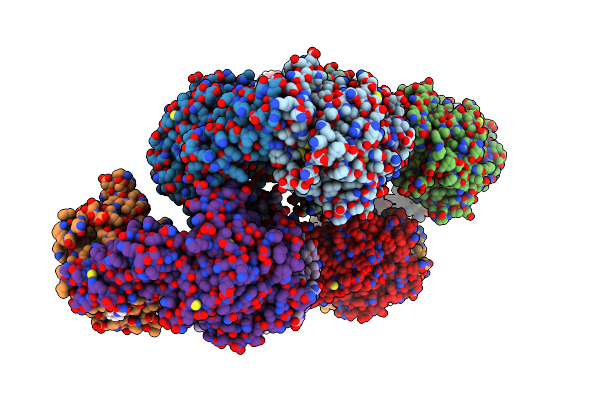
Crystal Structure Of The Quaternary Ammonium Rieske Monooxygenase Cnta In Complex With Substrate Gamma-Butyrobetaine
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2020-11-18 Classification: OXIDOREDUCTASE Ligands: FE, FES, NM2, GOL |
 |
Crystal Structure Of The Quaternary Ammonium Rieske Monooxygenase Cnta In Complex With Substrate L-Carnitine
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2020-11-18 Classification: OXIDOREDUCTASE Ligands: FE, FES, 152, SCN, EPE |
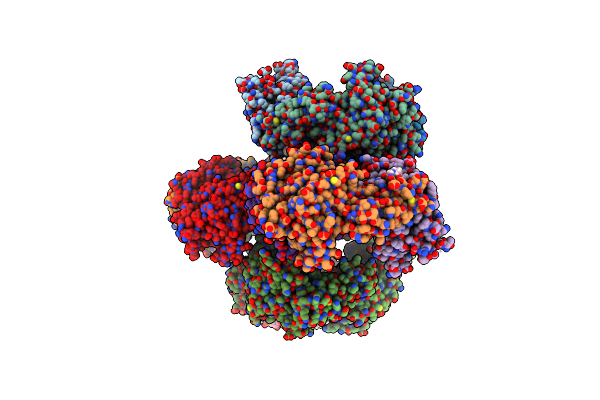 |
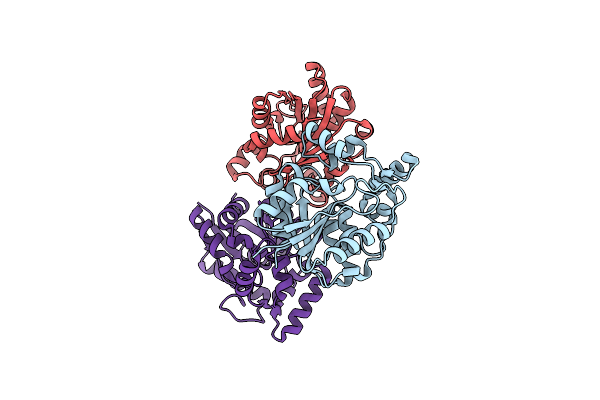
Crystal Structure Of The Quaternary Ammonium Rieske Monooxygenase Cnta In Complex With Inhibitor Mmv12 (Mmv020670)
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2020-11-18 Classification: OXIDOREDUCTASE Ligands: FE, FES, QKQ, EPE |
 |
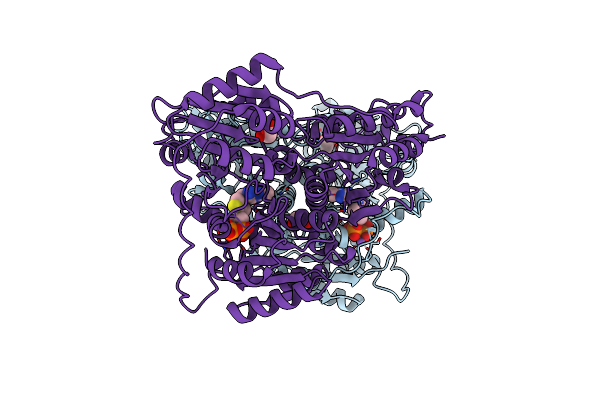
Extracellular Alpha/Beta-Hydrolase From Paenibacillus Species Shares Structural And Functional Homology To Tobacco Salicylic Acid Binding Protein 2
Organism: Paenibacillus sp. vtt e-133280
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2020-04-22 Classification: HYDROLASE |
 |
Pseudomonas Fluorescens Pf-5 Thiamine Diphosphate-Dependent 4-Hydroxybenzoylformate Decarboxylase
Organism: Pseudomonas protegens pf-5
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2020-01-08 Classification: OXIDOREDUCTASE Ligands: TPP, NA, EDO |
 |
Sphingobacterium Sp. T2 Manganese Superoxide Dismutase Catalyses The Oxidative Demethylation Of Polymeric Lignin Via Generation Of Hydroxyl Radical
Organism: Sphingobacterium spiritivorum
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2018-10-03 Classification: OXIDOREDUCTASE Ligands: MN |
 |
Sphingobacterium Sp. T2 Manganese Superoxide Dismutase Catalyses The Oxidative Demethylation Of Polymeric Lignin Via Generation Of Hydroxyl Radical
Organism: Sphingobacterium spiritivorum
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2018-10-03 Classification: OXIDOREDUCTASE Ligands: MN |
 |
Structural And Functional Characterisation Of A Bacterial Laccase-Like Multi-Copper Oxidase Cueo From Lignin-Degrading Bacterium Ochrobactrum Sp. With Oxidase Activity Towards Lignin Model Compounds And Lignosulfonate
Organism: Ochrobactrum
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2018-04-11 Classification: OXIDOREDUCTASE Ligands: CU, EDO |
 |
Structure Of Thermobifida Fusca Dyp-Type Peroxidase And Activity Towards Kraft Lignin And Lignin Model Compounds
Organism: Thermobifida fusca
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-04-06 Classification: OXIDOREDUCTASE Ligands: HEM, OXY |
 |
Organism: Sphingobacterium spiritivorum
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2015-08-05 Classification: OXIDOREDUCTASE Ligands: MN |
 |
Crystal Structure Of Yfau, A Metal Ion Dependent Class Ii Aldolase From Escherichia Coli K12
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2008-09-09 Classification: LYASE Ligands: PO4, GOL |
 |
Crystal Structure Of Yfau, A Metal Ion Dependent Class Ii Aldolase From Escherichia Coli K12 - Mg-Pyruvate Product Complex
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2008-09-09 Classification: LYASE Ligands: MG, PYR, PO4, GOL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2007-10-02 Classification: LYASE Ligands: PO4, GOL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2007-10-02 Classification: LYASE Ligands: MG, OXM, PO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2005-02-15 Classification: HYDROLASE Ligands: CL |

