Search Count: 48
 |
Pseudo-Atomic Model Of The Tetrahedral 24Mer Of Hsp17 From Caenorhabditis Elegans
Organism: Caenorhabditis elegans
Method: ELECTRON MICROSCOPY Release Date: 2022-12-14 Classification: CHAPERONE |
 |
Organism: Thermus parvatiensis
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2022-06-22 Classification: HYDROLASE Ligands: ZN, GOL, EPE, SO4, NA, CL |
 |
Crystal Structure Of The Human Hsp40 Dnajb1-Ctds In Complex With A Peptide Of Nudc
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2022-01-19 Classification: CHAPERONE Ligands: EDO |
 |
Pseudo-Atomic Model For Hsp26 Residues 63 To 214. Please Be Advised That The Target Map Is Not Of Sufficient Resolution To Unambiguously Position Backbone Or Side Chain Atoms. This Model Represents A Likely Fit.
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2021-11-24 Classification: CHAPERONE |
 |
Organism: Canis lupus familiaris, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2021-11-10 Classification: CHAPERONE |
 |
Organism: Canis lupus familiaris, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2021-11-10 Classification: CHAPERONE |
 |
Organism: Canis lupus familiaris, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2021-11-10 Classification: CHAPERONE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2020-03-18 Classification: IMMUNE SYSTEM Ligands: CA, PEG, PGE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-03-18 Classification: PROTEIN FIBRIL |
 |
Pseudo-Atomic Model Of A 16-Mer Assembly Of Reduced Recombinant Human Alphaa-Crystallin (Non Domain Swapped Configuration)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2019-12-11 Classification: CHAPERONE |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2019-11-20 Classification: UNKNOWN FUNCTION Ligands: GOL |
 |
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2019-11-20 Classification: UNKNOWN FUNCTION |
 |
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2019-11-20 Classification: VIRAL PROTEIN Ligands: GOL, ACT |
 |
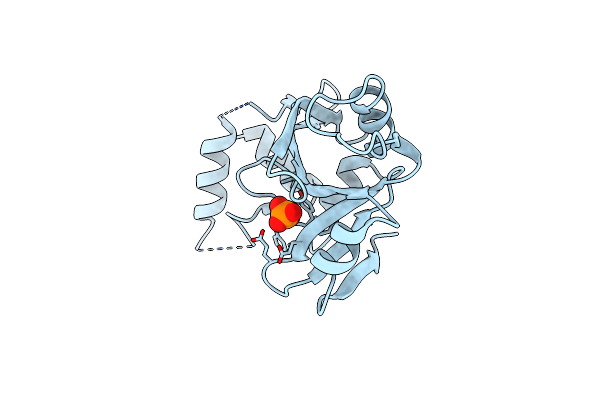
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-03-27 Classification: CHAPERONE Ligands: MG |
 |
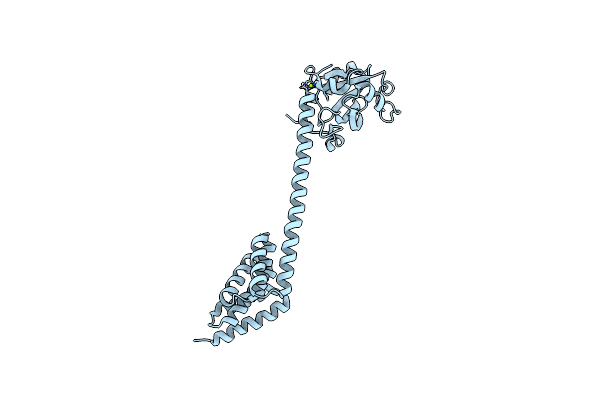
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2019-03-27 Classification: CHAPERONE Ligands: PO4 |
 |
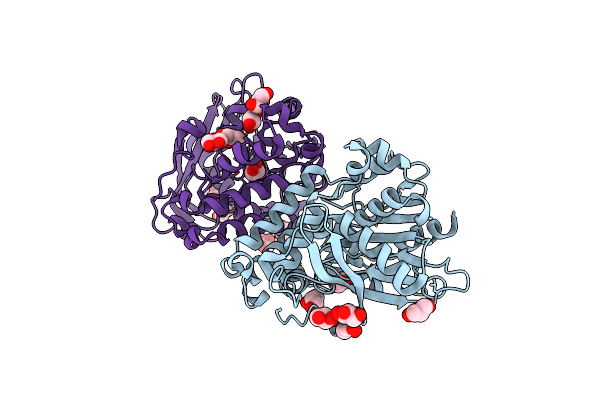
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2019-03-27 Classification: CHAPERONE Ligands: MG |
 |
Trigonal Crystal Structure Of An Acetylester Hydrolase From Corynebacterium Glutamicum
Organism: Corynebacterium glutamicum
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2015-12-16 Classification: HYDROLASE Ligands: GOL |
 |
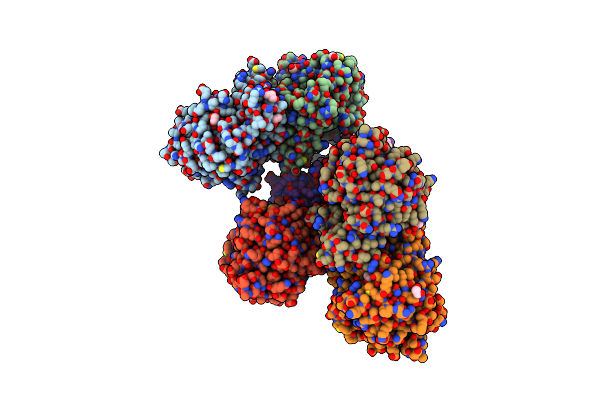
Organism: Pseudomonas veronii
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2015-12-16 Classification: HYDROLASE |
 |
Organism: Pseudomonas veronii
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2015-12-16 Classification: HYDROLASE Ligands: EDO, GOL, ACT |
 |
Orthorhombic Crystal Structure Of An Acetylester Hydrolase From Corynebacterium Glutamicum
Organism: Corynebacterium glutamicum (strain atcc 13032 / dsm 20300 / jcm 1318 / lmg 3730 / ncimb 10025)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-12-09 Classification: TRANSFERASE Ligands: CL, MG, GOL |

