Search Count: 18
 |
Cryo-Em Structure Of Mouse Tmem16F-Yfp Purified And Plunged Using Miso (Microfluidic Isolation)
Organism: Mus musculus, Aequorea victoria
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN Ligands: CA |
 |
E. Coli Beta-Galactosidase Labeled With Chromeo P503 Dye Purified Using Miso
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: HYDROLASE |
 |
E. Coli Beta-Galactosidase Labeled With Chromeo P503 Dye Purified Using Miso From 1Ug
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: HYDROLASE |
 |
Organism: Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN Ligands: NAG |
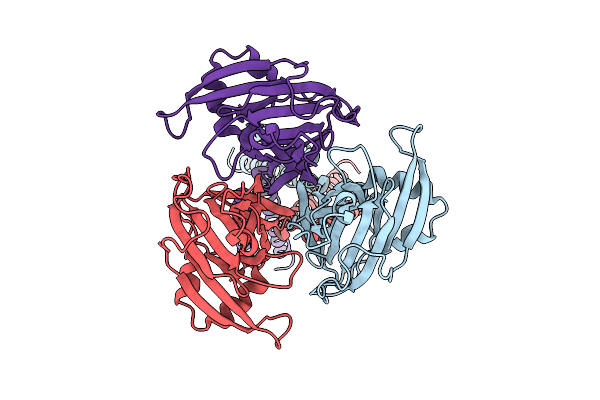 |
Cryo-Em Structure Of Bovine Tmem206-Yfp Purified And Plunged Using Miso (Micro-Purification)
Organism: Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN |
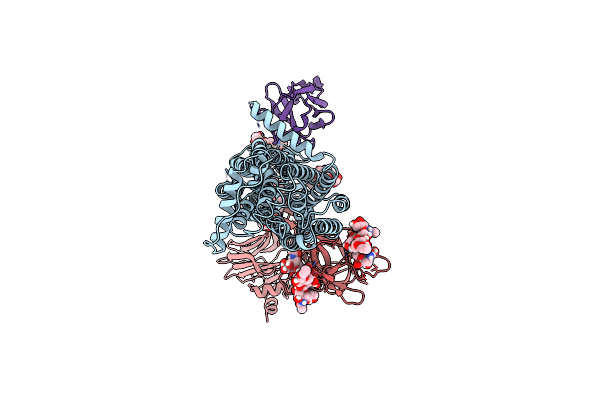 |
Cryo-Em Structure Of Native Sv2A In Complex With Tent-Hc, Gangliosides And Pro-Macrobody 5
Organism: Clostridium tetani e88, Lama glama, Escherichia coli (strain k12), Ovis aries
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: MEMBRANE PROTEIN Ligands: NAG, 6UZ, A1IHM, A1IHN |
 |
Cryo-Em Structure Of Native Sv2A In Complex With Tent-Hc, Pro-Macrobody 5 And Levetiracetam
Organism: Clostridium tetani e88, Lama glama, Escherichia coli k-12, Ovis aries
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: MEMBRANE PROTEIN Ligands: NAG, 6UZ, UKX |
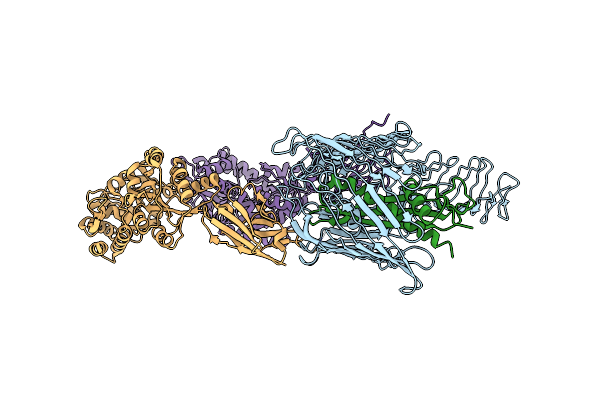 |
Cryo-Em Structure Of N. Gonorhoeae Lptde In Complex With Promacrobodies (Mbps Have Not Been Built De Novo)
Organism: Neisseria gonorrhoeae, Synthetic construct, Methanosarcina mazei
Method: ELECTRON MICROSCOPY Release Date: 2022-05-04 Classification: TRANSPORT PROTEIN |
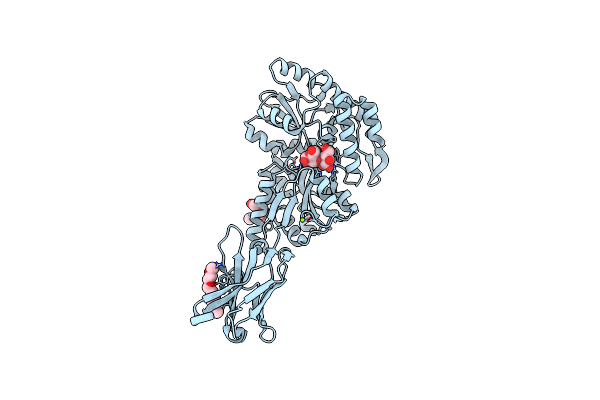 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-05-04 Classification: IMMUNE SYSTEM Ligands: P6G, MG |
 |
Crystal Structure Of The Lysosomal Potassium Channel Mttmem175 T38A Mutant Soaked With Zinc
Organism: Lama glama, Escherichia coli (strain k12), Marivirga tractuosa (strain atcc 23168 / dsm 4126 / nbrc 15989 / ncimb 1408 / vkm b-1430 / h-43)
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2020-04-15 Classification: TRANSPORT PROTEIN Ligands: LMT, K |
 |
Crystal Structure Of The Potassium Channel Mttmem175 In Complex With A Nanobody-Mbp Fusion Protein
Organism: Lama glama, Escherichia coli (strain k12), Marivirga tractuosa dsm 4126
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2019-08-28 Classification: TRANSPORT PROTEIN Ligands: LMT, K |
 |
Organism: Lama glama, Escherichia coli (strain k12), Marivirga tractuosa dsm 4126
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2019-08-28 Classification: TRANSPORT PROTEIN Ligands: LMT, RB |
 |
Organism: Lama glama, Escherichia coli (strain k12), Marivirga tractuosa dsm 4126
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2019-08-28 Classification: TRANSPORT PROTEIN Ligands: LMT, CS |
 |
Organism: Lama glama, Escherichia coli (strain k12), Marivirga tractuosa dsm 4126
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2019-08-28 Classification: TRANSPORT PROTEIN Ligands: LMT, K, ZN |
 |
Crystal Structure Of The Potassium Channel Mttmem175 T38A Variant In Complex With A Nanobody-Mbp Fusion Protein
Organism: Lama glama, Escherichia coli (strain k12), Marivirga tractuosa dsm 4126
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2019-08-28 Classification: TRANSPORT PROTEIN Ligands: LMT, K |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2017-06-07 Classification: MEMBRANE PROTEIN |
 |
Organism: Nectria haematococca
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2014-11-12 Classification: LIPID TRANSPORT Ligands: CA |
 |
Organism: Nectria haematococca
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2014-11-12 Classification: LIPID TRANSPORT Ligands: CA |

