Search Count: 72
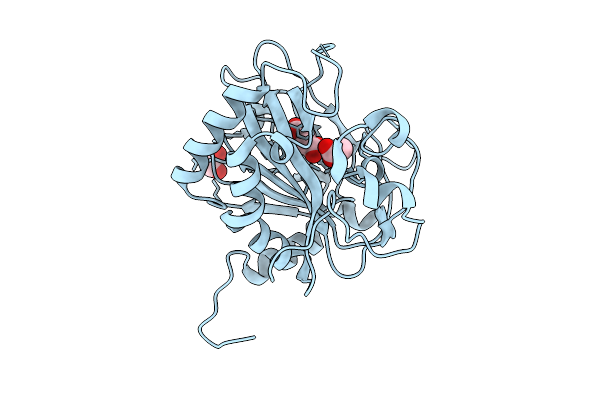 |
Crystal Structure Of Fe/2-Og Dependent Dioxygenase Mysh In Complex With Nickel And Succinate
Organism: Nostoc linckia nies-25
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: METAL BINDING PROTEIN Ligands: SIN, NI, ACT, EDO |
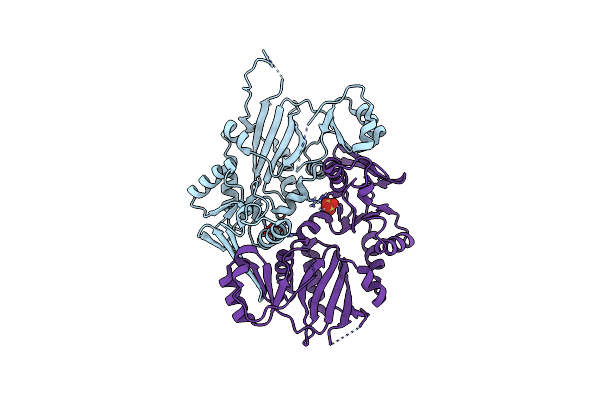 |
Crystal Structure Of Atp-Grasp Ligase Prub From Streptomyces Coelicolor A3(2)
Organism: Streptomyces coelicolor a3(2)
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2024-12-18 Classification: LIGASE Ligands: SO4 |
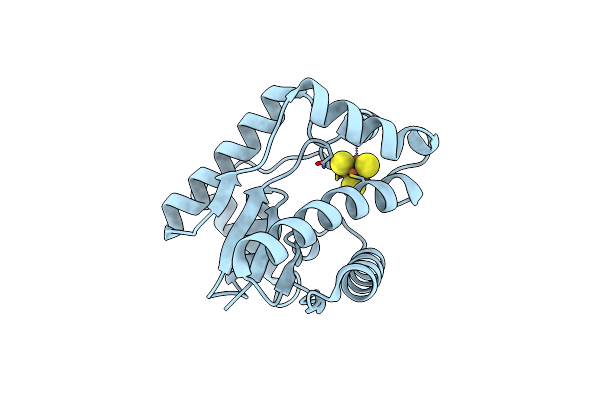 |
Crystal Structure Of Epoxyqueuosine Reductase Queh C9S Mutant From Thermotoga Maritima
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2024-12-18 Classification: METAL BINDING PROTEIN Ligands: SF4 |
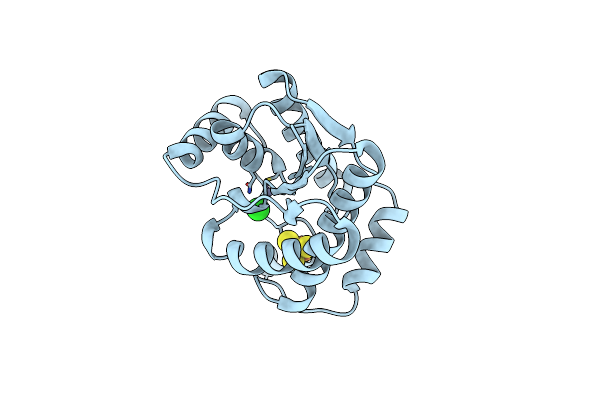 |
Crystal Structure Of Epoxyqueuosine Reductase Queh D13N Mutant From Thermotoga Maritima
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2024-12-18 Classification: METAL BINDING PROTEIN Ligands: SF4, ZN, CL |
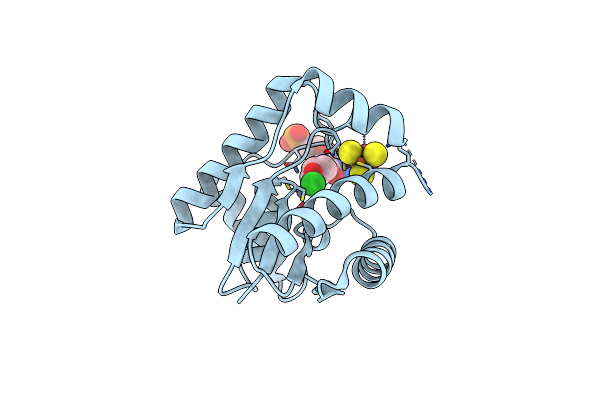 |
Crystal Structure Of Epoxyqueuosine Reductase Queh In Complex With Queuosine
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-12-18 Classification: METAL BINDING PROTEIN Ligands: 56B, SF4, CL, ZN |
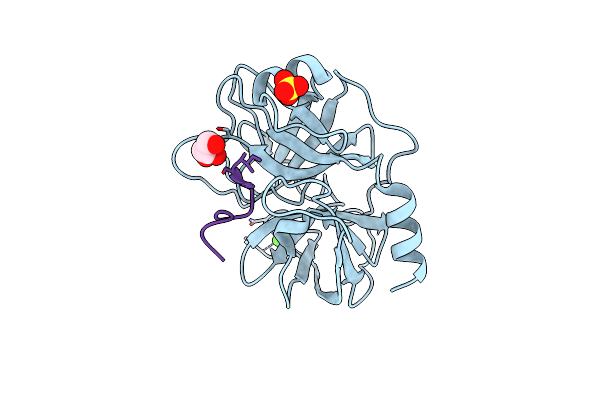 |
Organism: Microcystis aeruginosa mrc, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-02-14 Classification: HYDROLASE Ligands: SO4, CA, GOL |
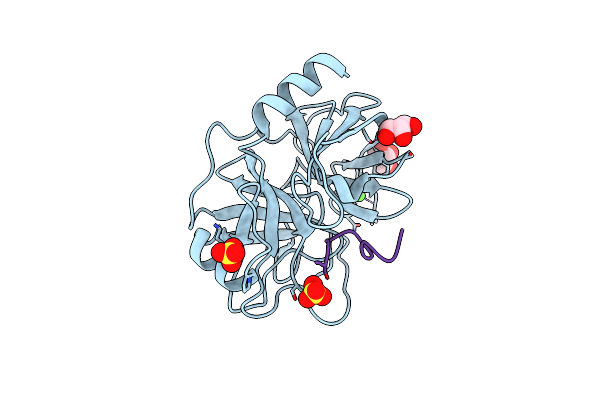 |
Organism: Microcystis aeruginosa mrc, Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2024-02-14 Classification: HYDROLASE Ligands: GOL, SO4, CA |
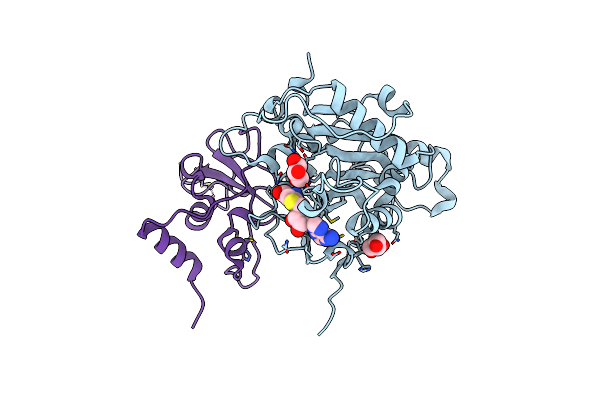 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-01-24 Classification: VIRAL PROTEIN Ligands: GOL, SAH, ZN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-09-20 Classification: BIOSYNTHETIC PROTEIN |
 |
The Structure Of Haemophilus Influenzae Rd Kw20 Nitroreductase Complexed With 1-Methyl-5-Nitroimidazole
Organism: Haemophilus influenzae rd kw20
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-02-23 Classification: OXIDOREDUCTASE Ligands: EDO, FMN, EIV, ACY |
 |
The Structure Of Haemophilus Influenzae Rd Kw20 Nitroreductase Complexed With Nicotinic Acid
Organism: Haemophilus influenzae rd kw20
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-02-23 Classification: OXIDOREDUCTASE Ligands: SO4, FMN, NIO |
 |
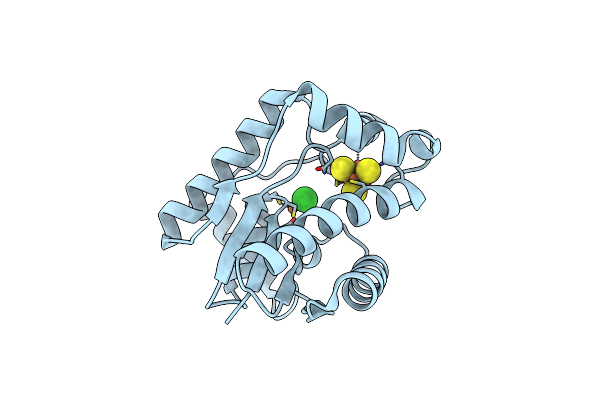
Crystal Structure Of Epoxyqueuosine Reductase Queh In Complex With Gmp From Thermotoga Maritima
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2022-01-12 Classification: OXIDOREDUCTASE Ligands: SF4, 5GP, FE, CL |
 |
Crystal Structure Of Epoxyqueuosine Reductase Queh From Thermotoga Maritima
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-11-03 Classification: OXIDOREDUCTASE Ligands: SF4, FE, CL |
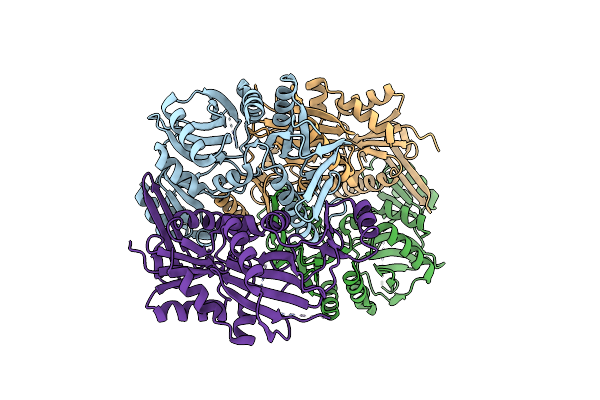 |
Organism: Nostoc sp. (strain pcc 7120 / sag 25.82 / utex 2576)
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2021-11-03 Classification: BIOSYNTHETIC PROTEIN |
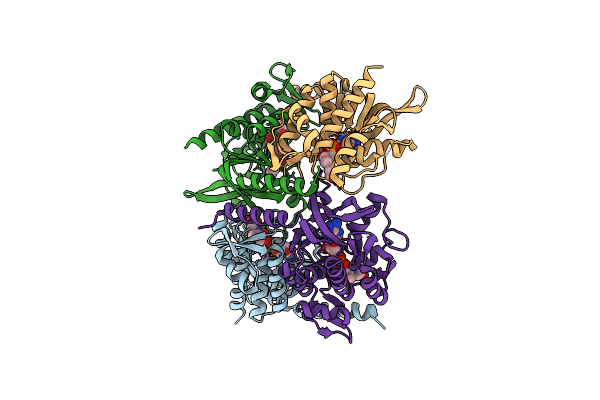 |
Organism: Thermovibrio ammonificans
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2021-09-01 Classification: BIOSYNTHETIC PROTEIN Ligands: FE, 48H |
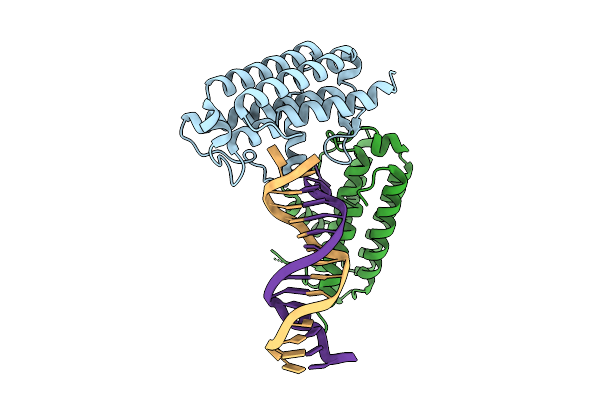 |
Crystal Structure Of Colibactin Self-Resistance Protein Clbs In Complex With A Dsdna
Organism: Escherichia coli, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2021-05-26 Classification: DNA BINDING PROTEIN/DNA |
 |
Crystal Structure Of Colibactin Self-Resistance Protein Clbs In Complex With Two Molecules Of Ches
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-05-26 Classification: DNA BINDING PROTEIN Ligands: NHE |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2021-02-17 Classification: LYASE Ligands: MN |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2020-09-30 Classification: LYASE Ligands: MN, GOL, CL, NA |
 |
Organism: Synechocystis sp. pcc 6803
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2020-03-04 Classification: LYASE |

