Search Count: 54
 |
Organism: Trichoderma pseudokoningii
Method: SOLUTION NMR Release Date: 2018-06-13 Classification: ANTIBIOTIC |
 |
Organism: Trichoderma pseudokoningii
Method: SOLUTION NMR Release Date: 2018-06-13 Classification: ANTIBIOTIC |
 |
Organism: Trichoderma pseudokoningii
Method: SOLUTION NMR Release Date: 2018-02-28 Classification: ANTIBIOTIC |
 |
Organism: Streptococcus pneumoniae
Method: SOLUTION NMR Release Date: 2017-05-10 Classification: HYDROLASE Ligands: CA |
 |
Solution Structure And Dynamics Of The Outer Membrane Cytochrome Omcf From Geobacter Sulfurreducens
Organism: Geobacter sulfurreducens (strain atcc 51573 / dsm 12127 / pca)
Method: SOLUTION NMR Release Date: 2017-04-12 Classification: ELECTRON TRANSPORT Ligands: HEC |
 |
Crystal Structure Of The Full-Length Cell Wall-Binding Module Of Cpl7 Mutant R223A
Organism: Streptococcus phage cp-7
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2017-03-08 Classification: HYDROLASE Ligands: GOL |
 |
Nmr Assignment And Structure Of A Peptide Derived From The Membrane Proximal External Region Of Hiv-1 Gp41 In The Presence Of Dodecylphosphocholine Micelles
Organism: Human immunodeficiency virus 1
Method: SOLUTION NMR Release Date: 2017-02-22 Classification: VIRAL PROTEIN |
 |
Nmr Assignment And Structure Of A Peptide Derived From The Membrane Proximal External Region Of Hiv-1 Gp41 In The Presence Of Hexafluoroisopropanol
Organism: Human immunodeficiency virus 1
Method: SOLUTION NMR Release Date: 2017-02-22 Classification: VIRAL PROTEIN |
 |
Last Common Ancestor Of Gram Negative Bacteria (Gnca) Class A Beta- Lactamase
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-01-18 Classification: HYDROLASE Ligands: SO4, GOL |
 |
Last Common Ancestor Of Gram-Negative Bacteria (Gnca4) Beta-Lactamase Class A
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2016-12-28 Classification: HYDROLASE Ligands: PG0, PEG |
 |
W229D And F290W Mutant Of The Last Common Ancestor Of Gram-Negative Bacteria (Gnca4) Beta-Lactamase Class A
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2016-12-21 Classification: HYDROLASE Ligands: GOL, EDO, PG0, PEG, PG4, PGE |
 |
W229D Mutant Of The Last Common Ancestor Of Gram-Negative Bacteria (Gnca) Beta-Lactamase Bound To 5(6)-Nitrobenzotriazole (Ts-Analog)
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2016-12-21 Classification: HYDROLASE Ligands: 6NT |
 |
W229D And F290W Mutant Of The Last Common Ancestor Of Gram-Negative Bacteria (Gnca4) Beta-Lactamase Class A Bound To 5(6)-Nitrobenzotriazole (Ts-Analog)
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2016-12-21 Classification: HYDROLASE Ligands: 6NT |
 |
Crystal Structure Of The Cysteine Desulfurase Csda From Escherichia Coli At 1.996 Angstroem Resolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-12-21 Classification: TRANSFERASE Ligands: PLP, GOL, CIT |
 |
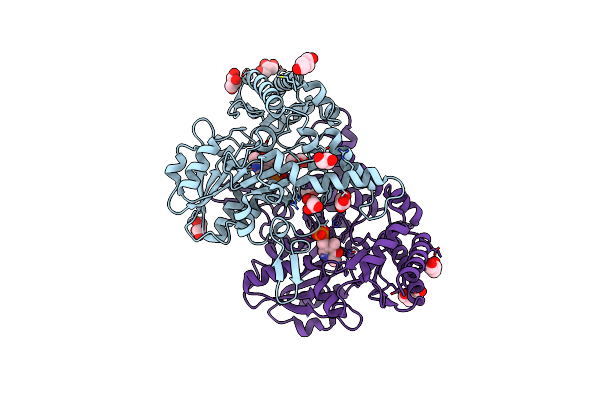
Crystal Structure Of The Cysteine Desulfurase Csda (Persulfurated) From Escherichia Coli At 2.384 Angstroem Resolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2016-11-23 Classification: TRANSFERASE Ligands: GOL, PEG, TLA, PLP |
 |
Crystal Structure Of The Cysteine Desulfurase Csda (S-Sulfonic Acid) From Escherichia Coli At 2.050 Angstroem Resolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2016-11-23 Classification: TRANSFERASE Ligands: GOL, PEG, PLP |
 |
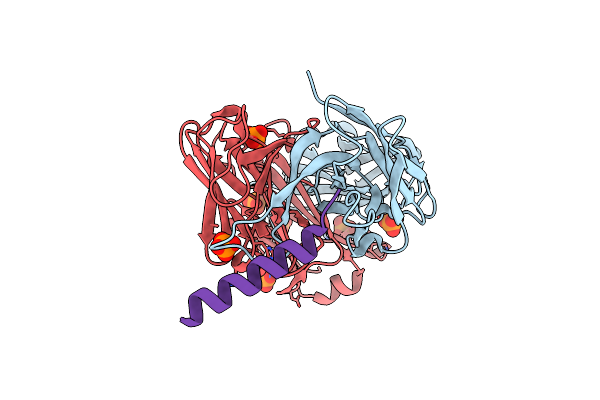
Crystal Structure Of The Complex Between The Cysteine Desulfurase Csda And The Sulfur-Acceptor Csde In The Persulfurated State At 2.50 Angstroem Resolution
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-11-23 Classification: TRANSFERASE Ligands: PLP, PEG, GOL |
 |
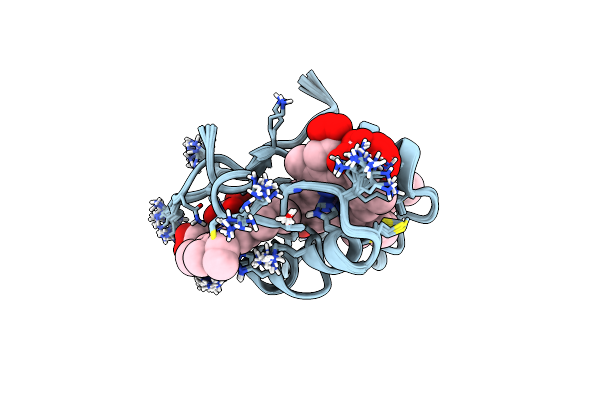
Crystal Structure Of Broad Neutralizing Antibody 10E8 With Long Epitope Bound
Organism: Homo sapiens, Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-11-23 Classification: IMMUNE SYSTEM Ligands: PO4 |
 |
A Key Amino Acid In The Control Of Different Functional Behavior Within The Triheme Cytochrome Family From Geobacter Sulfurreducens
Organism: Geobacter sulfurreducens
Method: SOLUTION NMR Release Date: 2016-09-21 Classification: ELECTRON TRANSPORT Ligands: HEM |
 |
Molecular Basis For Protein Recognition Specificity Of The Dynlt1/Tctex1 Canonical Binding Groove. Characterization Of The Interaction With Activin Receptor Iib
|

