Search Count: 12
 |
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2011-06-01 Classification: OXIDOREDUCTASE Ligands: FAD, CL, SAR |
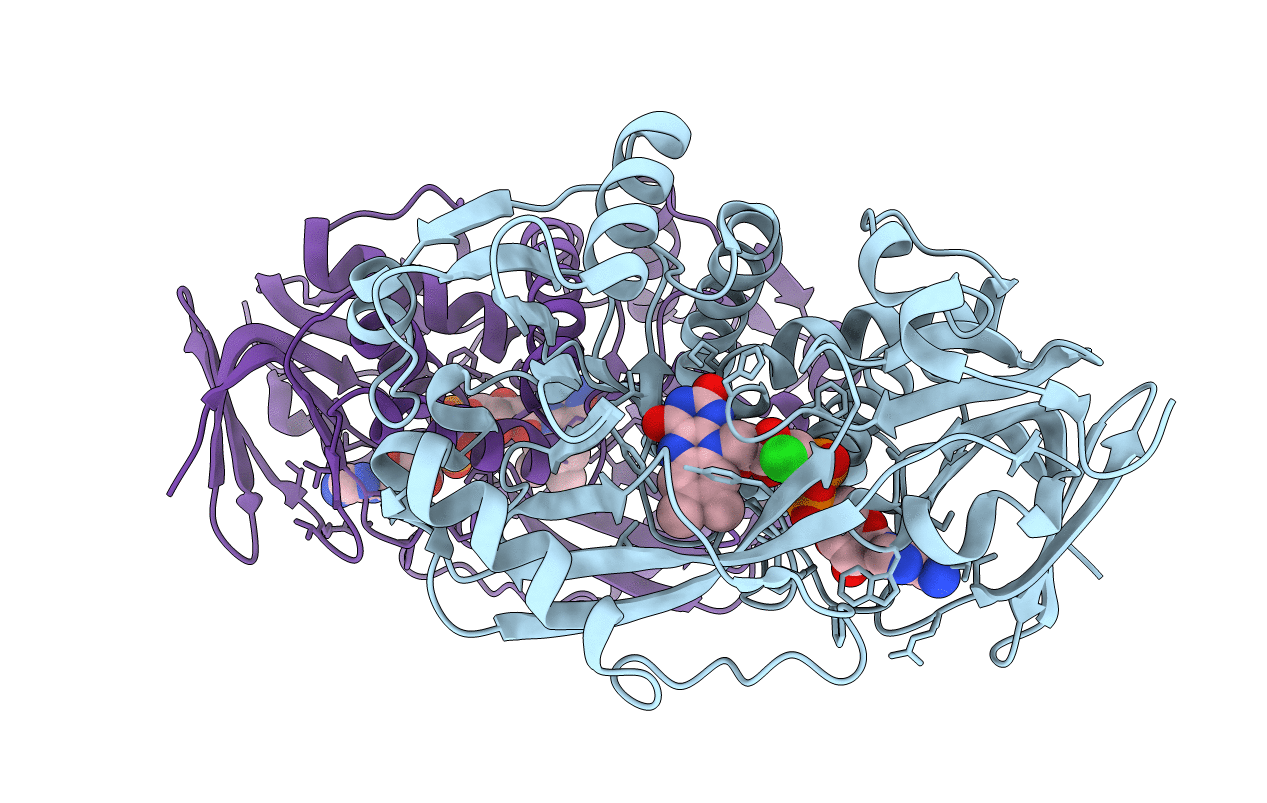 |
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-06-01 Classification: OXIDOREDUCTASE Ligands: FAD, CL |
 |
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2011-06-01 Classification: OXIDOREDUCTASE Ligands: FAD, CL, MTG |
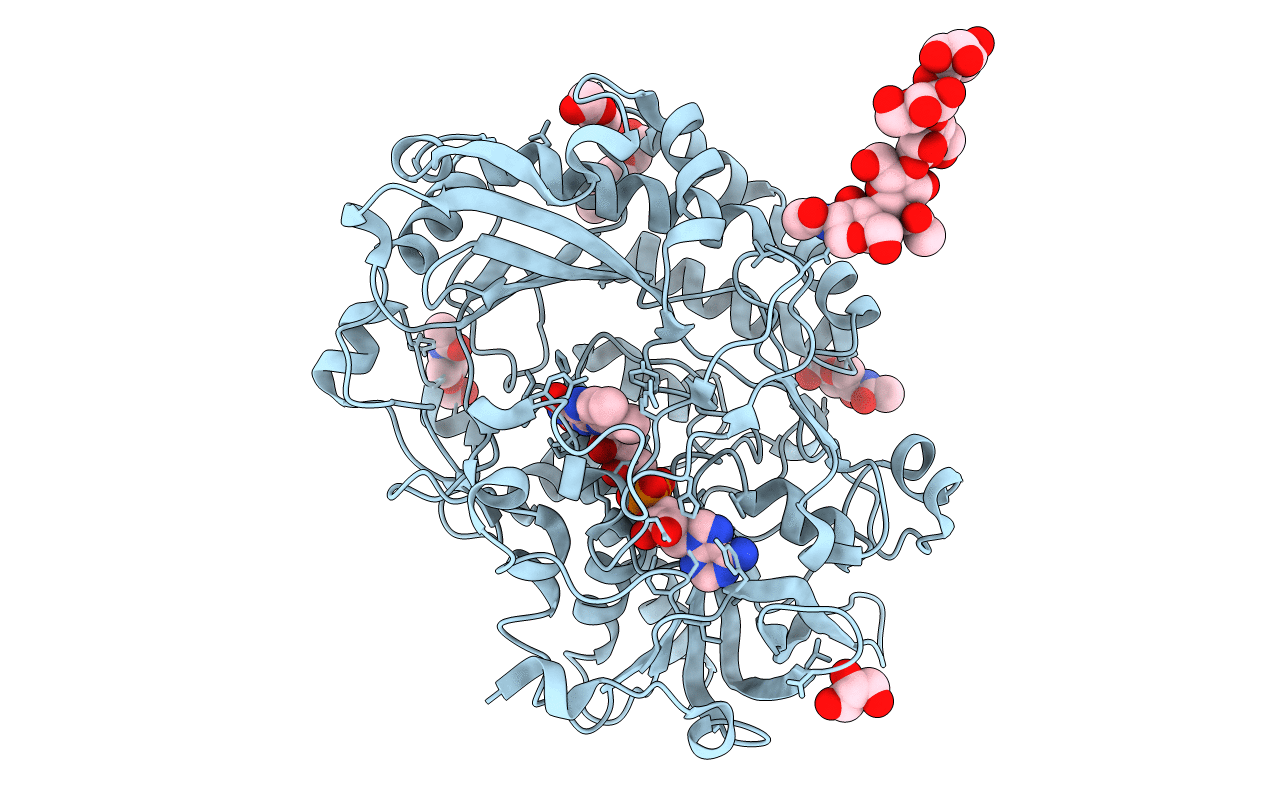 |
Crystal Structure Of Glucose Oxidase For Space Group C2221 At 1.2 A Resolution
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2011-06-01 Classification: OXIDOREDUCTASE Ligands: NAG, FAD, GOL, PEG, CL |
 |
Crystal Structure Of Glucose Oxidase For Space Group P3121 At 1.3 A Resolution.
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2011-06-01 Classification: OXIDOREDUCTASE Ligands: NAG, FAD, CL |
 |
Trypsin In Complex With The Inhibitor 1-[3-(Aminomethyl)Phenyl]-N-[3-Fluoro-2'-(Methylsulfonyl)Biphenyl-4-Yl]-3-(Trifluoromethyl)-1H-Pyrazole-5-Carboxamide (Dpc423)
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-04-21 Classification: HYDROLASE Ligands: M35, CA |
 |
Factor Xa In Complex With The Inhibitor 1-[3-(Aminomethyl)Phenyl]-N-[3-Fluoro-2'-(Methylsulfonyl)Biphenyl-4-Yl]-3-(Trifluoromethyl)-1H-Pyrazole-5-Carboxamide (Dpc423)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2010-04-21 Classification: HYDROLASE Ligands: M35, CA |
 |
Factor Xa In Complex With The Inhibitor 1-[2-(Aminomethyl)Phenyl]-N-(3-Fluoro-2'-Sulfamoylbiphenyl-4-Yl)-3-(Trifluoromethyl)-1H-Pyrazole-5-Carboxamide (Dpc602)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2010-04-21 Classification: HYDROLASE Ligands: M37, CA |
 |
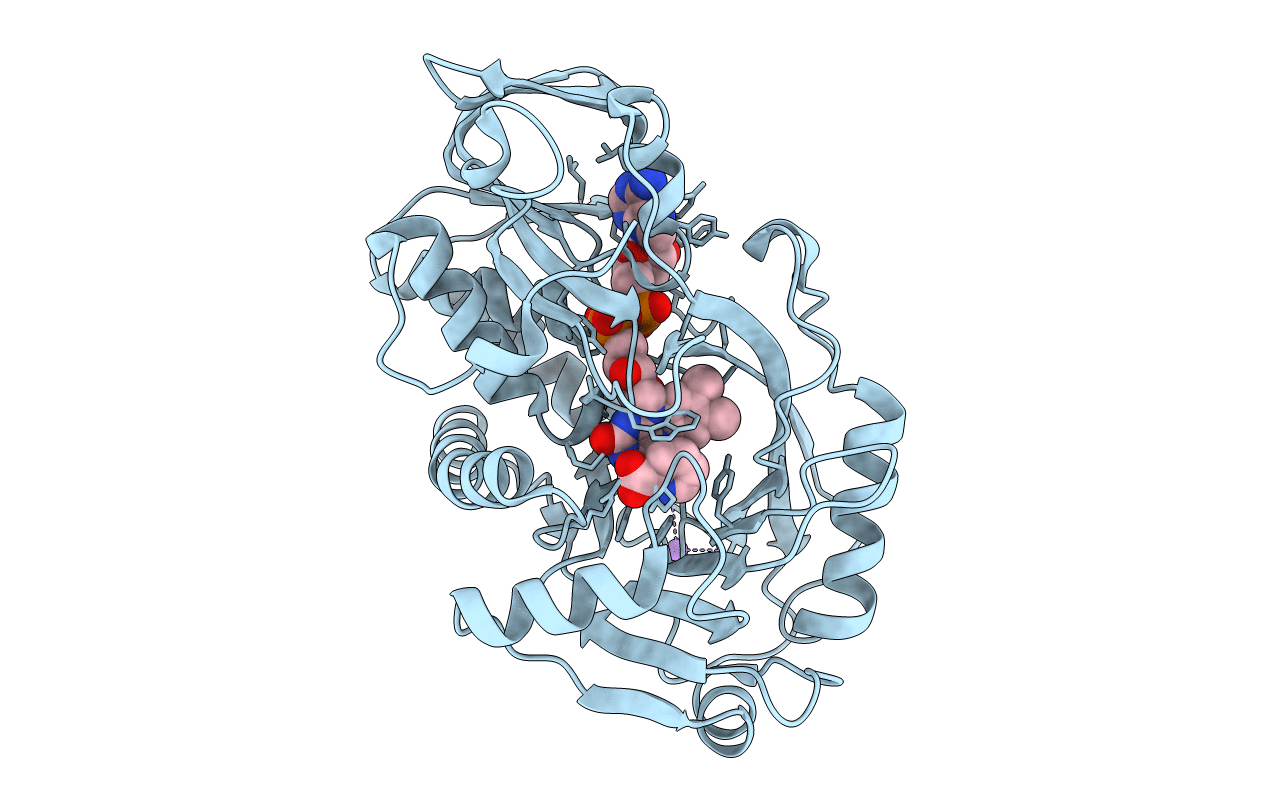
Tyr258Phe Mutant Of Nikd, An Unusual Amino Acid Oxidase Essential For Nikkomycin Biosynthesis: Open Form At 1.55A Resolution
Organism: Streptomyces tendae
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2009-10-20 Classification: OXIDOREDUCTASE Ligands: FAD, 6PC, MPD |
 |
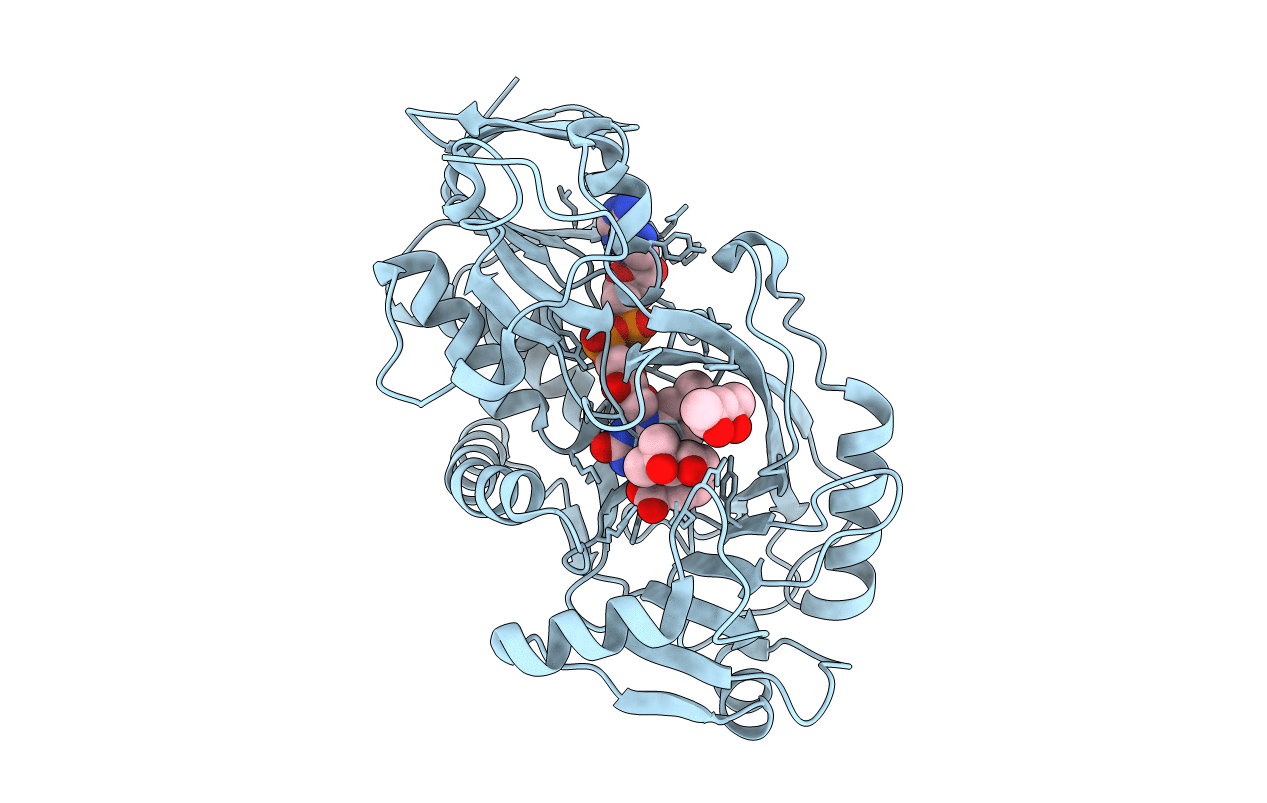
Nikd, An Unusual Amino Acid Oxidase Essential For Nikkomycin Biosynthesis: Closed Form At 1.15 A Resolution
Organism: Streptomyces tendae
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2007-07-31 Classification: OXIDOREDUCTASE |
 |
Nikd, An Unusual Amino Acid Oxidase Essential For Nikkomycin Biosynthesis: Open Form At 1.9A Resolution
Organism: Streptomyces tendae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2007-07-31 Classification: OXIDOREDUCTASE Ligands: FAD, 6PC, MPD |
 |
Organism: Streptomyces tendae
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2007-07-31 Classification: FLAVOPROTEIN Ligands: FAD, BEZ |

