Search Count: 18
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2022-09-28 Classification: BIOSYNTHETIC PROTEIN Ligands: PO4 |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-09-28 Classification: BIOSYNTHETIC PROTEIN |
 |
Crystal Structure Of Evolved Photoenzyme Ent1.3 (Truncated) With Bound Product
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-09-28 Classification: BIOSYNTHETIC PROTEIN Ligands: JRP, EDO |
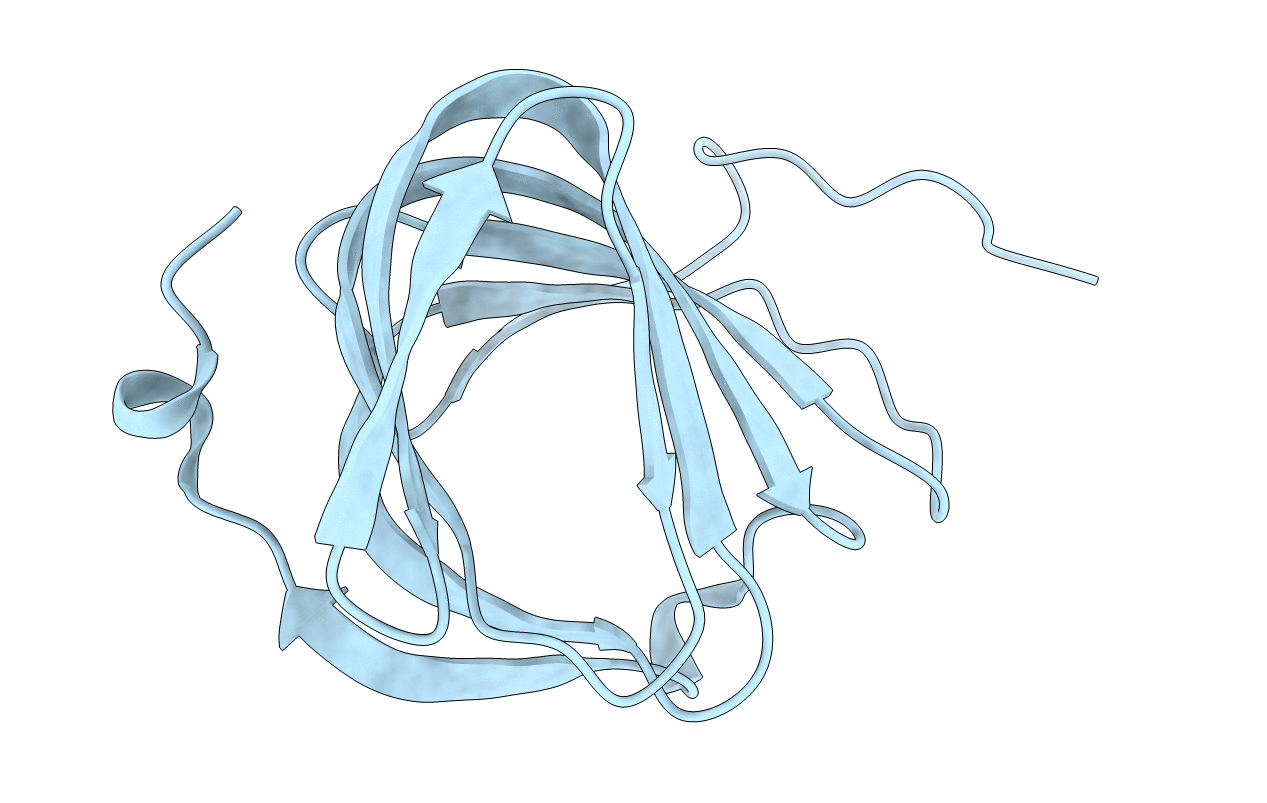 |
Roll Out The Beta-Barrel: Structure And Mechanism Of Pac13, A Unique Nucleoside Dehydratase
Organism: Streptomyces coeruleorubidus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2018-03-28 Classification: STRUCTURAL PROTEIN |
 |
Roll Out The Beta-Barrel: Structure And Mechanism Of Pac13, A Unique Nucleoside Dehydratase
Organism: Streptomyces coeruleorubidus
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2018-03-28 Classification: STRUCTURAL PROTEIN |
 |
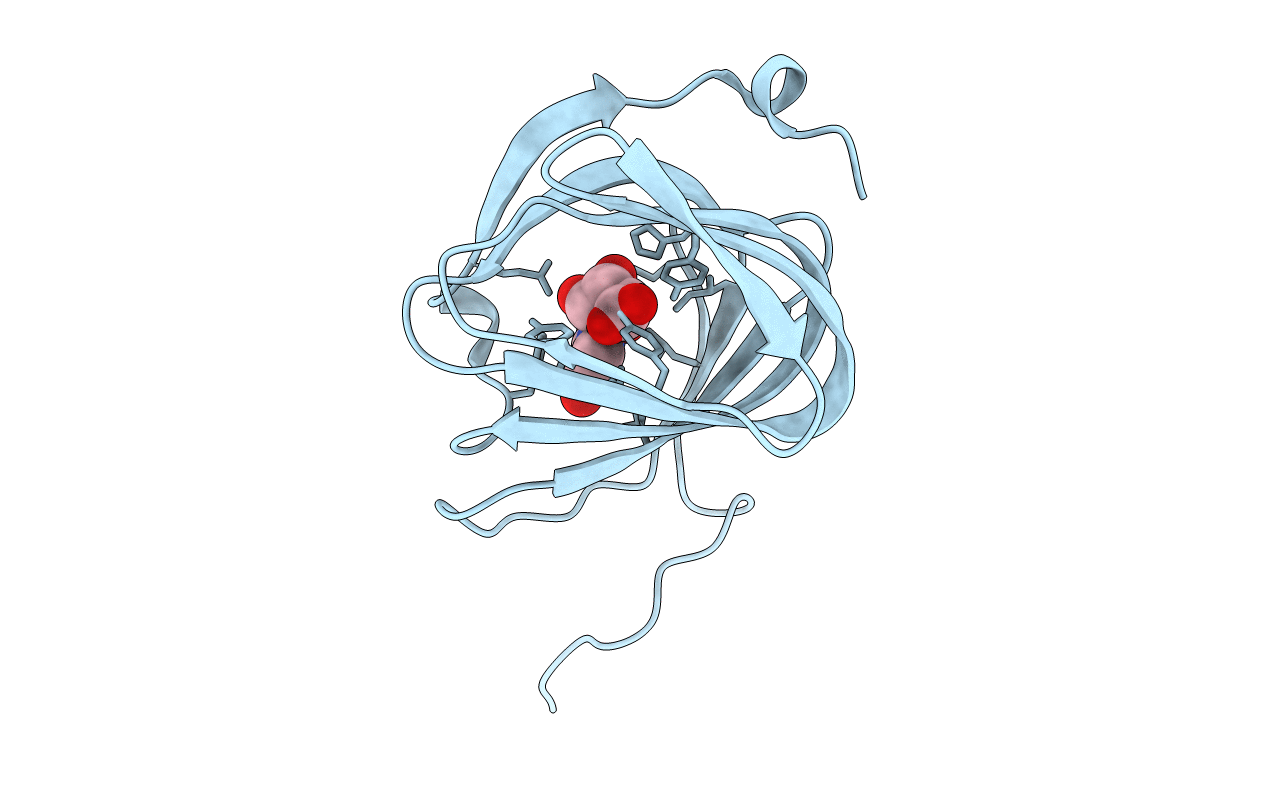
Organism: Streptomyces coeruleorubidus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-08-23 Classification: BIOSYNTHETIC PROTEIN Ligands: URI |
 |
Organism: Streptomyces coeruleorubidus
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2017-08-23 Classification: BIOSYNTHETIC PROTEIN Ligands: UUA |
 |
Organism: Streptomyces coeruleorubidus
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2017-08-23 Classification: BIOSYNTHETIC PROTEIN Ligands: EDO, URI |
 |
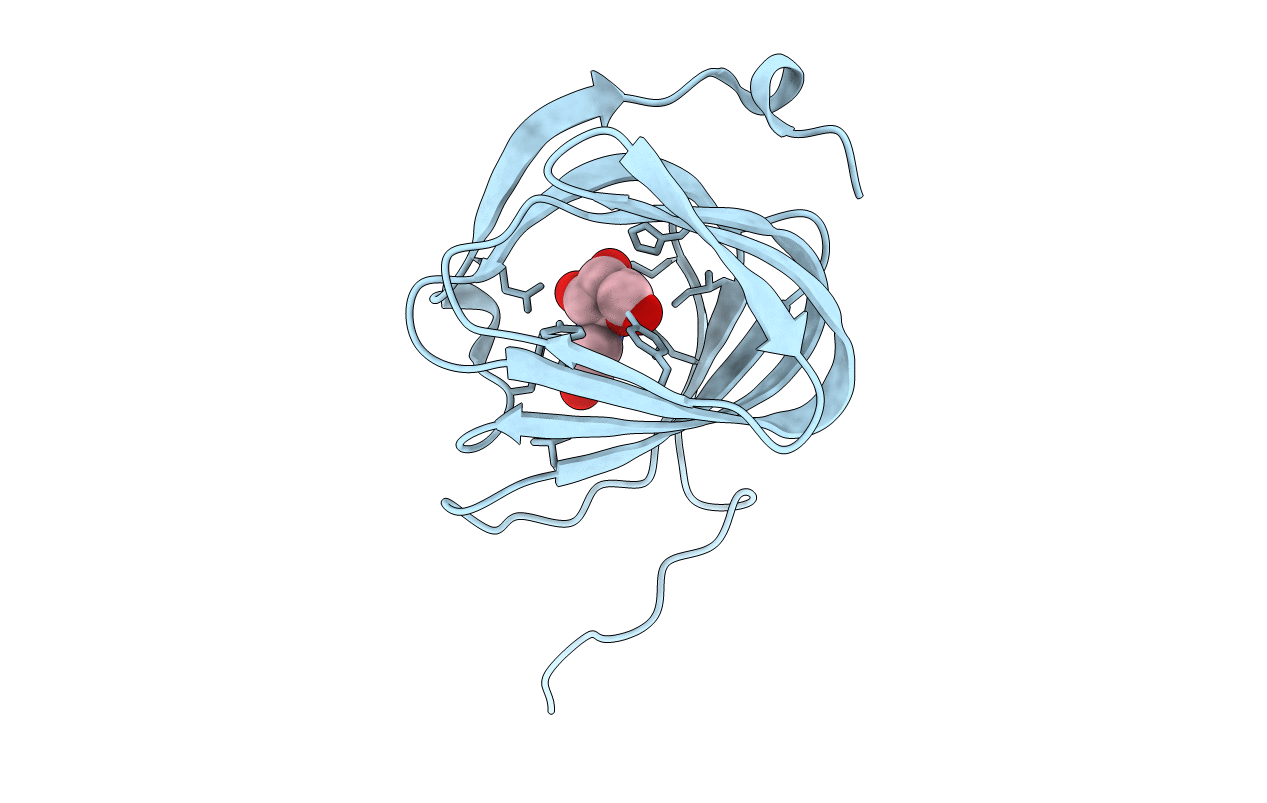
Organism: Streptomyces coeruleorubidus
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2017-08-23 Classification: BIOSYNTHETIC PROTEIN Ligands: URI |
 |
Organism: Streptomyces coeruleorubidus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-08-23 Classification: BIOSYNTHETIC PROTEIN Ligands: URI |
 |
Crystal Structure Of Anthrax Protective Antigen K446M Mutant To 1.91-A Resolution
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2012-10-24 Classification: TOXIN, TRANSPORT PROTEIN Ligands: CA |
 |
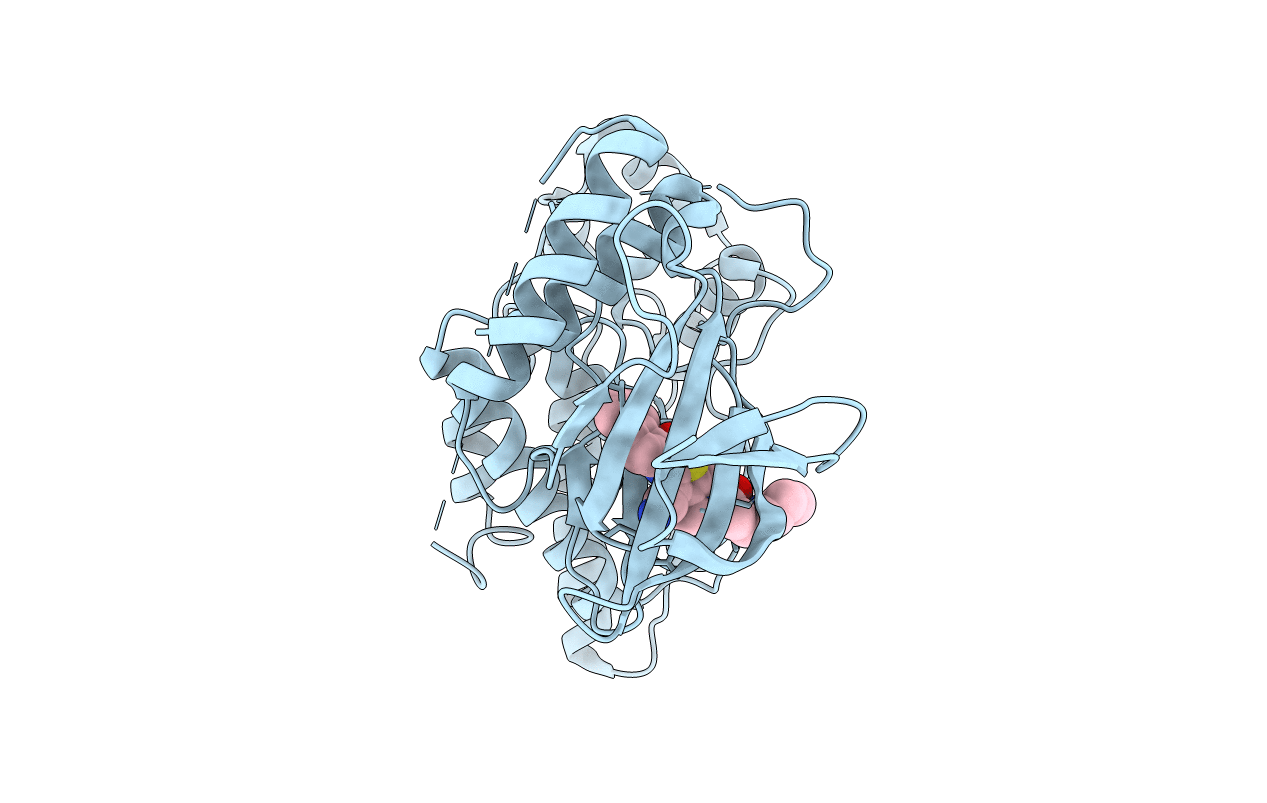
Crystal Structure Of Human Jnk3 Complexed With A 1-Aryl-3,4- Dihydroisoquinoline Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2009-03-31 Classification: TRANSFERASE Ligands: SNB |
 |
Crystal Structure Of Human Jnk3 Complexed With N-{3-Cyano-6-[3-(1-Piperidinyl)Propanoyl]-4,5,6,7-Tetrahydrothieno[2,3-C]Pyridin-2-Yl}-1-Naphthalenecarboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2007-02-27 Classification: TRANSFERASE Ligands: C0M |
 |
Crystal Structure Of Human Jnk3 Complexed With N-(3-Cyano-4,5,6,7-Tetrahydro-1-Benzothien-2-Yl)-2-Fluorobenzamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2007-02-27 Classification: TRANSFERASE Ligands: 738 |
 |
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2006-09-05 Classification: HYDROLASE/DNA Ligands: CA |
 |
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2006-09-05 Classification: HYDROLASE/DNA Ligands: CA |
 |
Co-Crystal Structure Of Checkpoint Kinase Chk1 With A Pyrrolo-Pyridine Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2006-06-13 Classification: SIGNALING PROTEIN, TRANSFERASE Ligands: SO4, 199 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2002-10-03 Classification: TRANSFERASE Ligands: SO4, TRS |

