Search Count: 114
 |
Crystal Structure Of Irak4-Hsa Complexed With N-[(2R)-2-Fluoro-3-Hydroxy-3-Methylbutyl]-6-[(5-Fluoropyri-Yl)Amino]-4-[(Propan-2-Yl)Amino]Pyridine-3-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2025-02-12 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: ZVD, SO4 |
 |
Crystal Structure Of Irak4-Hsa Complexed With Bms-986126; 6-((5-Cyano-2-Pyrimidinyl)Amino)-N-((2R)-2-Fluoro-3-Hydroxhylbutyl)-4-(Isopropylamino)Nicotinamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2025-02-12 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: ZVG, SO4 |
 |
Crystal Structure Of Irak4-Hsa Complexed With Bms-986147; 6-{5-Cyano-1H-Pyrazolo[3,4-B]Pyridin-1-Yl}-N-[(2R)-2-Fluorroxy-3-Methylbutyl]-4-[(Propan-2-Yl)Amino]Pyridine-3-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2025-02-12 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: SO4, ZVA |
 |
Crystal Structure Of Fluoroacetate Dehalogenase Daro3835 H274N Mutant With D107-Glycolyl Intermediate
Organism: Dechloromonas aromatica rcb
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-02-14 Classification: HYDROLASE |
 |
Organism: Dechloromonas aromatica rcb
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2023-09-06 Classification: HYDROLASE Ligands: CL |
 |
Organism: Saccharomyces cerevisiae, Xenopus laevis, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-06-07 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
 |
Organism: Xenopus laevis, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-06-07 Classification: DNA BINDING PROTEIN/DNA |
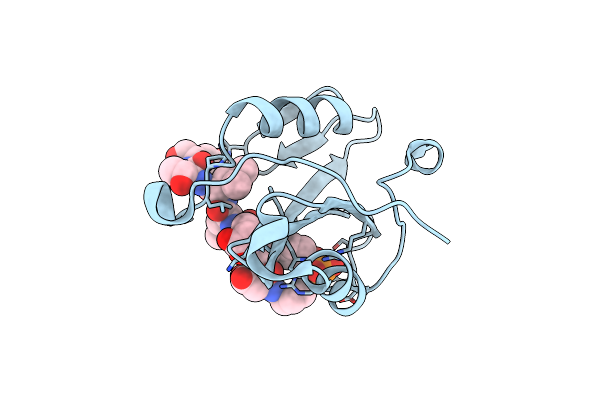 |
Crystal Structure Of The E372K Lnk Sh2 Domain Mutant In Complex With A Jak2 Py813 Phosphopeptide
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-05-31 Classification: SIGNALING PROTEIN Ligands: CL |
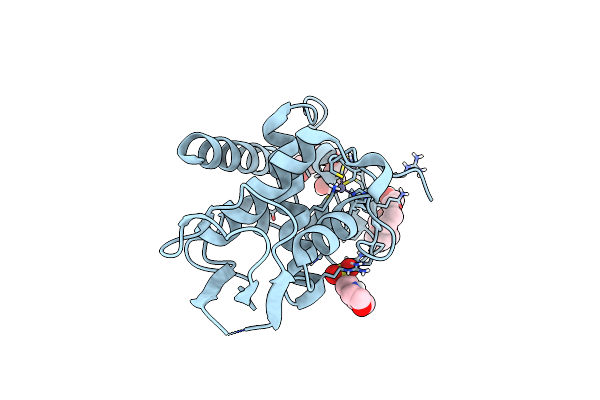 |
Organism: Vanderwaltozyma polyspora dsm 70294
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-03-29 Classification: DNA BINDING PROTEIN Ligands: ZN, MES, 1PE, PG4 |
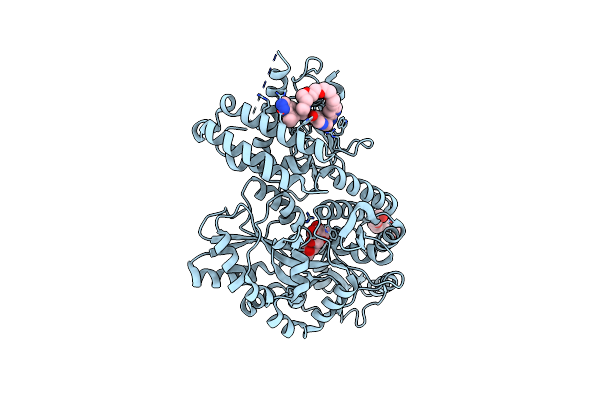 |
Crystal Structure Of A Cgrp Receptor Ectodomain Heterodimer Bound To Macrocyclic Inhibitor Htl0028125
Organism: Escherichia coli (strain k12), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-06-15 Classification: MEMBRANE PROTEIN Ligands: 7IR, PG4 |
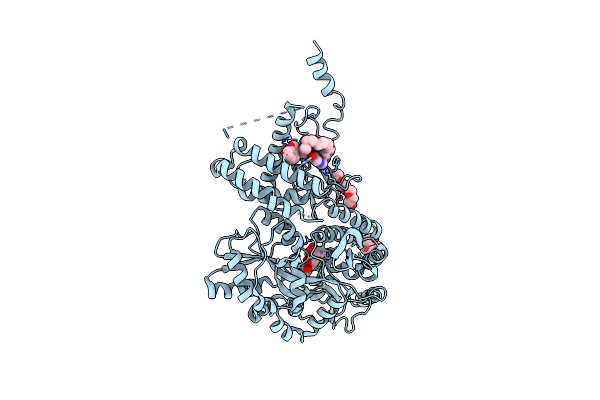 |
Crystal Structure Of A Cgrp Receptor Ectodomain Heterodimer Bound To Macrocyclic Inhibitor Compound 13
Organism: Escherichia coli (strain k12), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-06-15 Classification: MEMBRANE PROTEIN Ligands: PG4, 7IU |
 |
Organism: Homo sapiens, Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2021-10-06 Classification: MEMBRANE PROTEIN Ligands: QJT, OLA, PO4, OLC, EPE, PGE |
 |
Organism: Homo sapiens, Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2021-10-06 Classification: MEMBRANE PROTEIN Ligands: QK8, OLA, PGE, PO4 |
 |
Organism: Homo sapiens, Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-10-06 Classification: MEMBRANE PROTEIN Ligands: QK2, EPE, OLA, PO4 |
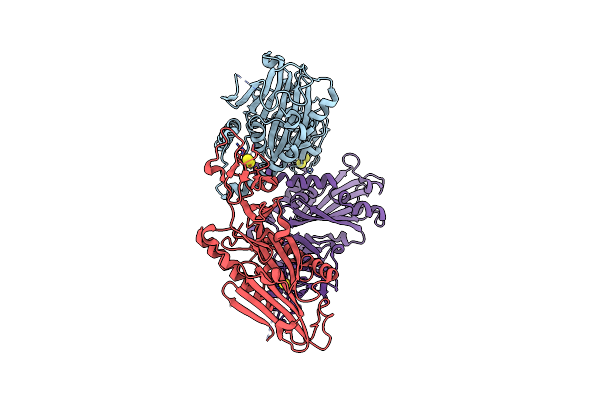 |
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2020-02-26 Classification: OXIDOREDUCTASE Ligands: FES |
 |
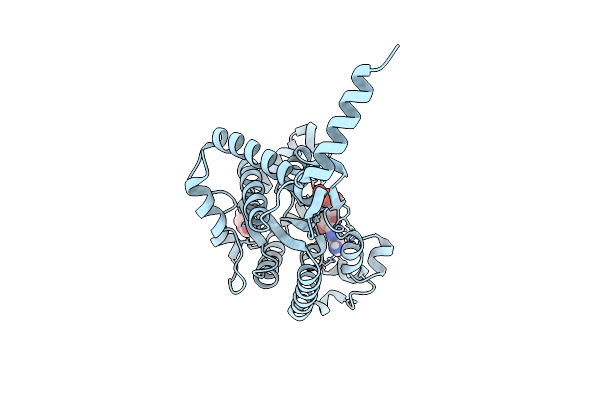
Crystal Structure Of The A2A-Star2-Bril562 In Complex With Azd4635 At 2.0A Resolution
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-06-26 Classification: MEMBRANE PROTEIN |
 |
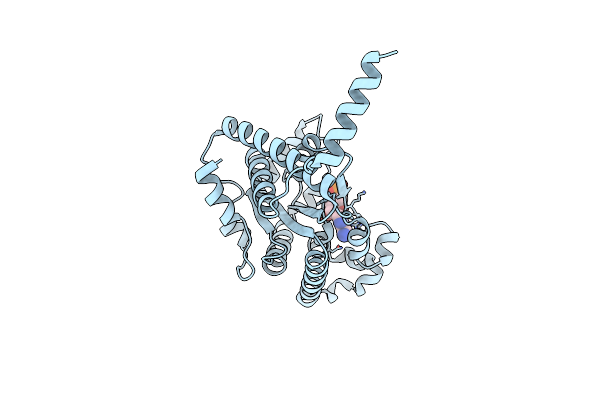
Crystal Structure Of Ppk2 Class Iii In Complex With Adp From Cytophaga Hutchinsonii Atcc 33406
Organism: Cytophaga hutchinsonii (strain atcc 33406 / ncimb 9469)
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2019-01-16 Classification: TRANSFERASE Ligands: ADP, GOL |
 |
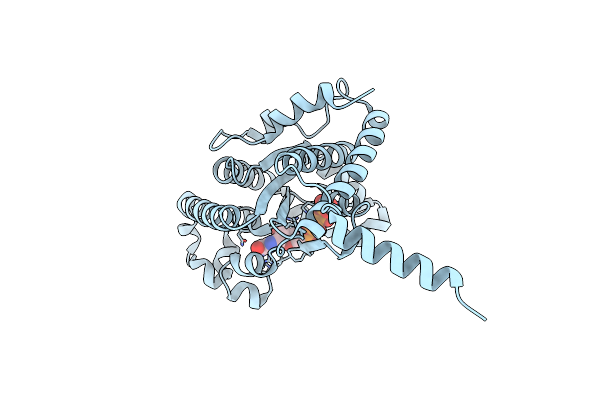
Crystal Structure Of Ppk2 Class Iii In The Complex With Amp From Cytophaga Hutchinsonii Atcc 33406
Organism: Cytophaga hutchinsonii (strain atcc 33406 / ncimb 9469)
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2019-01-16 Classification: TRANSFERASE Ligands: AMP, CL |
 |
Crystal Structure Of Ppk2 Class Iii In Complex With Guanosine 5-Tetraphosphate
Organism: Cytophaga hutchinsonii (strain atcc 33406 / ncimb 9469)
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2019-01-16 Classification: TRANSFERASE Ligands: BKP |
 |
Organism: Deinococcus radiodurans (strain atcc 13939 / dsm 20539 / jcm 16871 / lmg 4051 / nbrc 15346 / ncimb 9279 / r1 / vkm b-1422)
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2019-01-16 Classification: TRANSFERASE Ligands: ATP, MG, GOL, MPD, CL |

