Search Count: 64
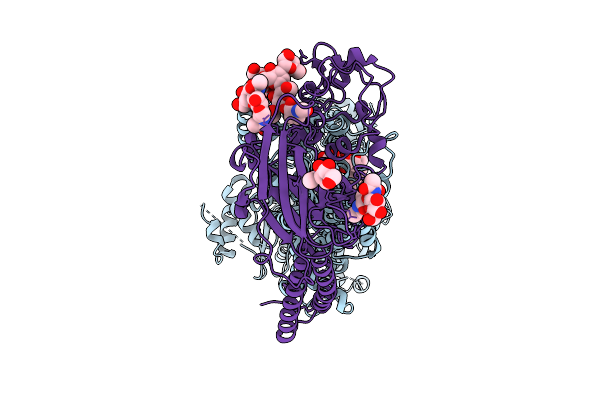 |
Cryo-Em Structure Of The C. Neoformans Lipid Flippase Apt1-Cdc50 Bound With Butyrolactol A In The E2P State
Organism: Cryptococcus neoformans var. grubii h99
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: TRANSLOCASE/INHIBITOR Ligands: NAG, BEF, MG, A1BD6 |
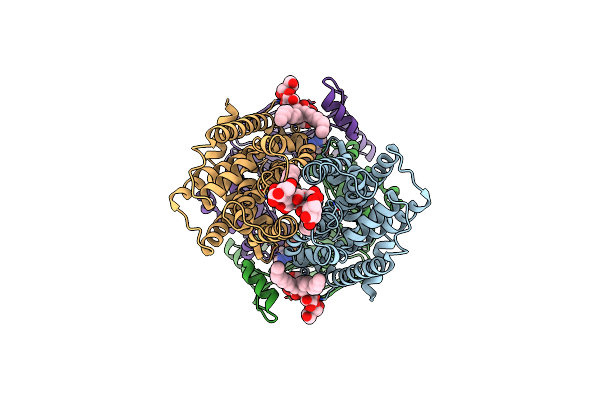 |
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: MEMBRANE PROTEIN Ligands: AV0, AGS, MG |
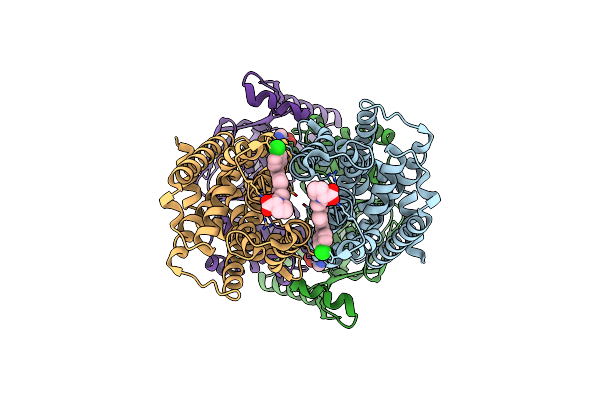 |
Cryo-Em Structure Of S. Aureus Targh In Complex With Amp-Pnp And Targocil-Ii
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: MEMBRANE PROTEIN Ligands: A1AV9, ANP, MG |
 |
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: MEMBRANE PROTEIN Ligands: AV0, AGS, MG |
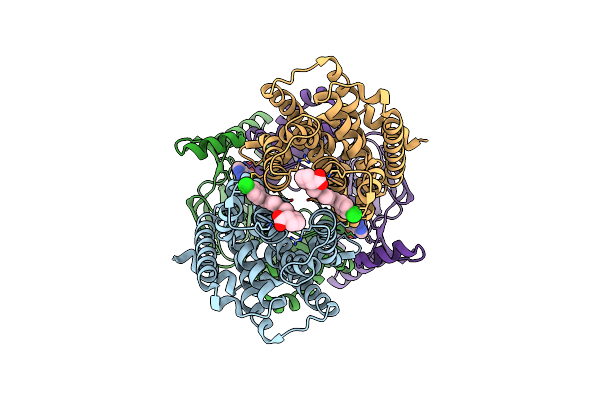 |
Cryo-Em Structure Of S. Aureus Targh In Complex With Targocil-Ii And Atp-Gamma-S In A Catalytically Competent Conformation
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: MEMBRANE PROTEIN Ligands: A1AV9, MG, AGS |
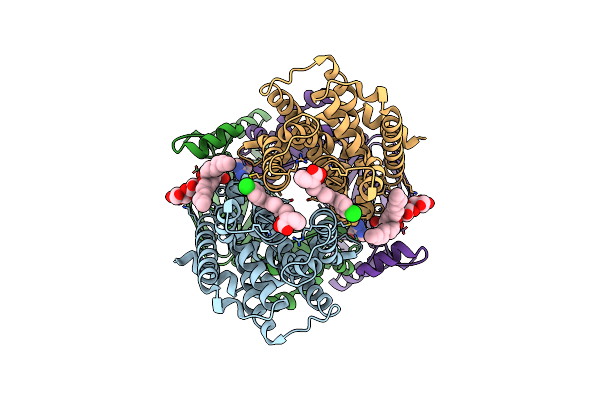 |
Cryo-Em Structure Of S. Aureus Targh In Complex With Targocil-Ii And Atp-Gamma-S In A Catalytically Incompetent Conformation
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: MEMBRANE PROTEIN Ligands: AV0, A1AV9, AGS, MG |
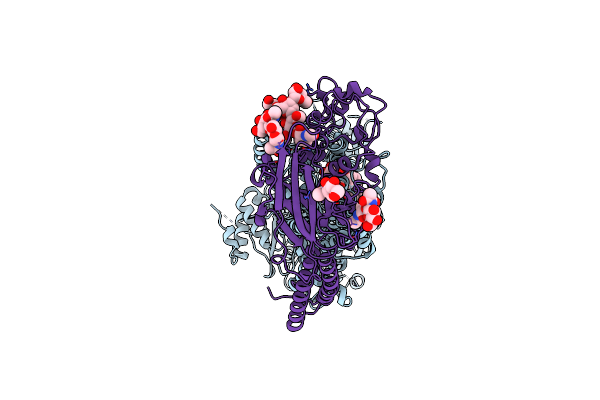 |
Cryo-Em Structure Of The C. Neoformans Lipid Flippase Apt1-Cdc50 Bound With Butyrolactol A In The E2P State
Organism: Cryptococcus neoformans var. grubii h99
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: TRANSLOCASE/INHIBITOR Ligands: NAG, BEF, MG, A1BD6 |
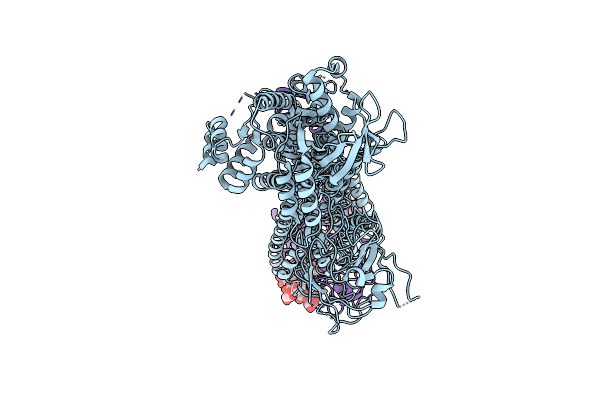 |
Cryo-Em Structure Of The C. Neoformans Lipid Flippase Apt1-Cdc50 In The E1 State
Organism: Cryptococcus neoformans var. grubii h99
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: LIPID TRANSPORT Ligands: NAG, MG |
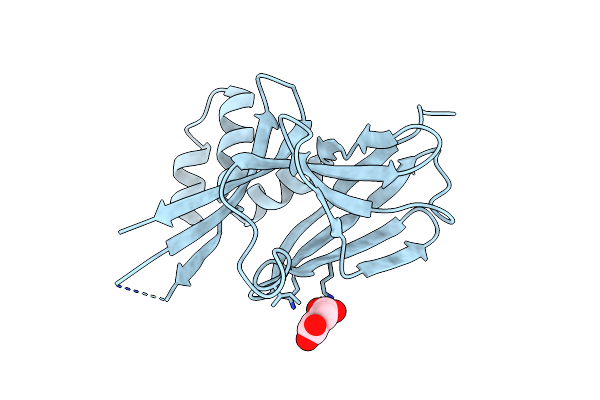 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-04-03 Classification: TRANSFERASE Ligands: SIN |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2024-04-03 Classification: TRANSFERASE |
 |
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2024-04-03 Classification: TRANSFERASE Ligands: V2V |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-08-04 Classification: TRANSFERASE Ligands: PEG |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2021-08-04 Classification: TRANSFERASE Ligands: 53Q |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-08-04 Classification: TRANSFERASE Ligands: CIT, V07 |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-08-04 Classification: TRANSFERASE Ligands: V0D |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2021-04-21 Classification: OXIDOREDUCTASE |
 |
Organism: Staphylococcus aureus (strain nctc 8325)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2021-04-21 Classification: OXIDOREDUCTASE Ligands: NDP |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-04-21 Classification: TRANSFERASE Ligands: PG4 |
 |
Crystal Structure Of S. Aureus Tari In Complex With Cdp-Ribitol (Space Group C121)
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2021-04-21 Classification: TRANSFERASE Ligands: V2V |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-04-21 Classification: TRANSFERASE Ligands: SCN |

