Search Count: 19
 |
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2020-07-22 Classification: HYDROLASE Ligands: ACT, SO4 |
 |
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2020-07-22 Classification: HYDROLASE Ligands: ACT, SO4 |
 |
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2020-07-22 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2020-07-22 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2020-07-22 Classification: HYDROLASE Ligands: ACT |
 |
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2020-07-22 Classification: HYDROLASE Ligands: ACT, SO4 |
 |
Crystal Structure Of Kemp Eliminase Hg3 With Bound Transition State Analogue, 277K
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2020-07-22 Classification: HYDROLASE Ligands: 6NT, SO4 |
 |
Crystal Structure Of Kemp Eliminase Hg3.3B With Bound Transition State Analogue, 277K
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2020-07-22 Classification: HYDROLASE Ligands: 6NT, SO4 |
 |
Crystal Structure Of Kemp Eliminase Hg3.7 With Bound Transition State Analogue, 277K
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2020-07-22 Classification: HYDROLASE Ligands: 6NT, SO4 |
 |
Crystal Structure Of Kemp Eliminase Hg3.14 With Bound Transition State Analogue, 277K
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-07-22 Classification: HYDROLASE Ligands: 6NT, SO4 |
 |
Crystal Structure Of Kemp Eliminase Hg3.17 With Bound Transition State Analog, 277K
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2020-07-22 Classification: HYDROLASE Ligands: 6NT |
 |
Crystal Structure Of Kemp Eliminase Hg4 With Bound Transition State Analogue, 277K
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2020-07-22 Classification: HYDROLASE Ligands: 6NT, SO4 |
 |
Solution Nmr Structure Of Dancer3-F34A, A Rigid And Natively Folded Single Mutant Of The Dynamic Protein Dancer-3
Organism: Streptococcus sp. group g
Method: SOLUTION NMR Release Date: 2019-08-21 Classification: DE NOVO PROTEIN |
 |
Structure Of The H477R Variant Of Rat Cytosolic Pepck In Complex With Manganese.
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-04-12 Classification: LYASE Ligands: MN, NA |
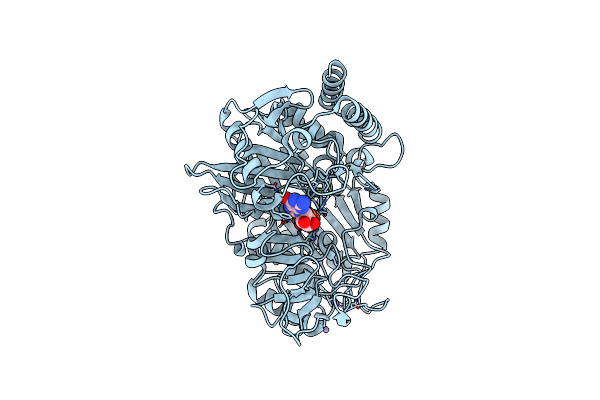 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-04-12 Classification: LYASE Ligands: MN, NA, GTP |
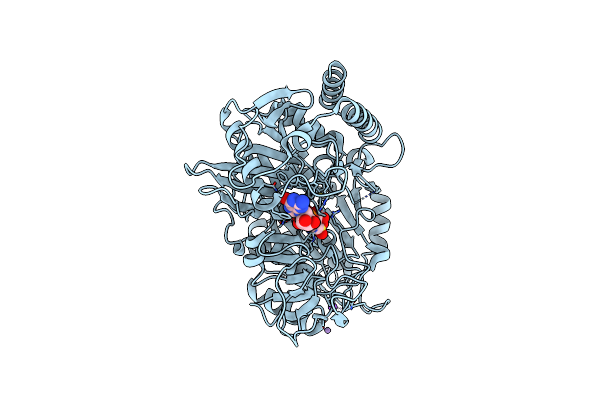 |
Structure Of The H477R Variant Of Rat Cytosolic Pepck In Complex With Beta Sulfopyruvate And Gtp.
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2017-04-12 Classification: LYASE Ligands: MN, NA, GTP, SPV |
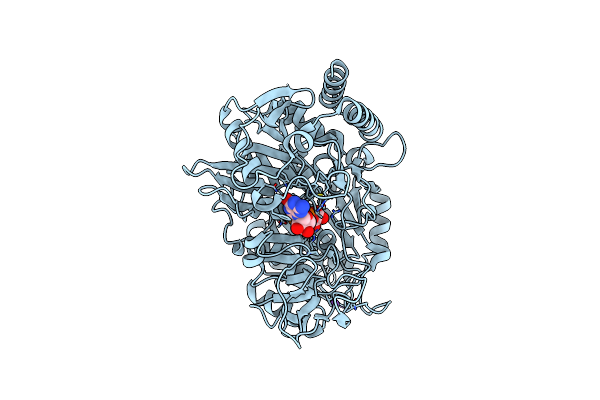 |
Structure Of The H477R Variant Of Rat Cytosolic Pepck In Complex With Oxalate And Gtp.
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-04-12 Classification: LYASE Ligands: MN, GTP, OXL, NA |
 |
Structure Of The H477R Variant Of Rat Cytosolic Pepck In Complex With Phosphoglycolate And Gdp.
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2017-04-12 Classification: LYASE Ligands: 1PE, MN, NA, GDP, PGA |
 |
Organism: Artificial gene
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2011-12-21 Classification: DE NOVO PROTEIN Ligands: BTB, GOL, NA |

