Search Count: 7
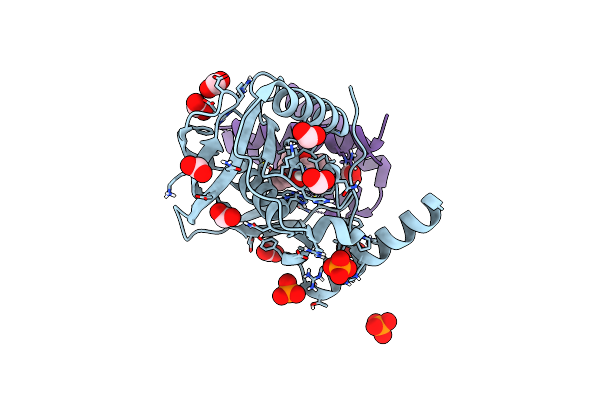 |
Crystal Structure Of Actinonin-Bound Pdf1 And The Computationally Designed Dbact553_1 Protein Binder
Organism: Pseudomonas aeruginosa, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2024-10-30 Classification: DE NOVO PROTEIN Ligands: ZN, BB2, FMT, PO4, K |
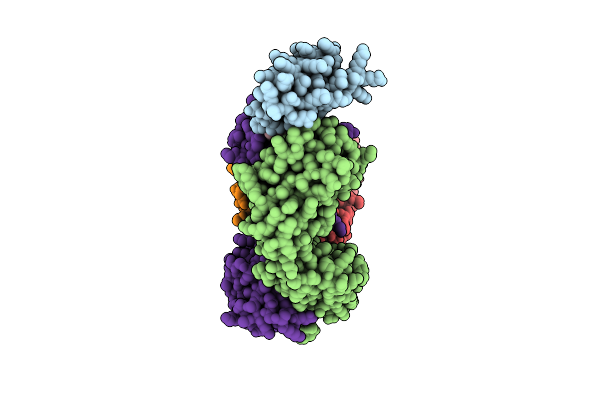 |
Progesterone-Bound Db3 Fab In Complex With Computationally Designed Dbpro1156_2 Protein Binder
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: DE NOVO PROTEIN Ligands: STR |
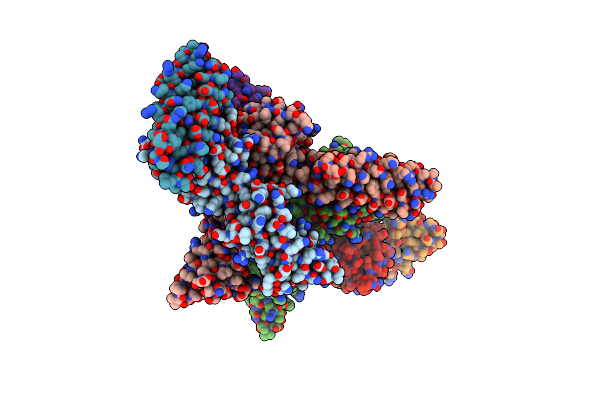 |
Organism: Homo sapiens, Chemical production metagenome
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2023-04-12 Classification: IMMUNE SYSTEM |
 |
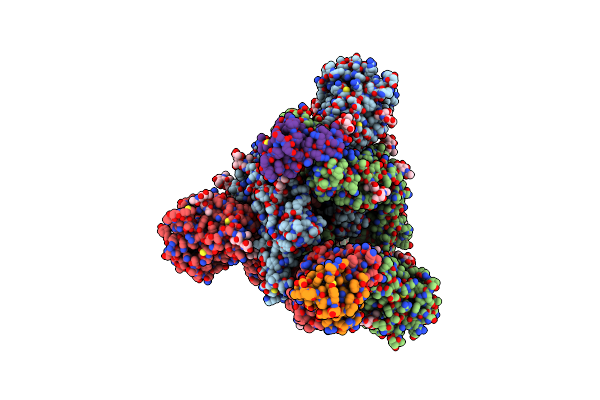
Crystal Strucutre Of Pd-L1 And The Computationally Designed Dbl1_03 Protein Binder
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2023-04-12 Classification: IMMUNE SYSTEM Ligands: ARG |
 |
Cryo-Em Structure Of Omicron Spike In Complex With De Novo Designed Binder, Full Map
Organism: Severe acute respiratory syndrome coronavirus 2, Tequatrovirus t4, Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2023-03-08 Classification: ANTIVIRAL PROTEIN Ligands: NAG |
 |
Cryo-Em Structure Of Omicron Spike In Complex With De Novo Designed Binder, Local
Organism: Severe acute respiratory syndrome coronavirus 2, Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2023-03-01 Classification: ANTIVIRAL PROTEIN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2023-03-01 Classification: ANTIVIRAL PROTEIN Ligands: NAG |

