Search Count: 9
 |
Mecp2 Is A Microsatellite Binding Protein That Protects Ca Repeats From Nucleosome Invasion
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-06-16 Classification: DNA BINDING PROTEIN |
 |
Nmr Structure Of Uhrf1 Tandem Tudor Domains In A Complex With Histone H3 Peptide
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2011-04-13 Classification: DNA BINDING PROTEIN/GENE REGULATION |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2010-08-18 Classification: MOTOR PROTEIN Ligands: MG, ADP |
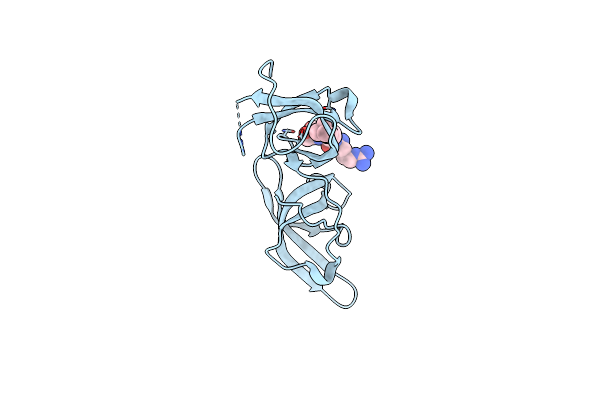 |
Crystal Structure Of The Tandem Tudor Domains Of The E3 Ubiquitin-Protein Ligase Uhrf1 In Complex With Trimethylated Histone H3-K9 Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2008-09-16 Classification: LIGASE |
 |
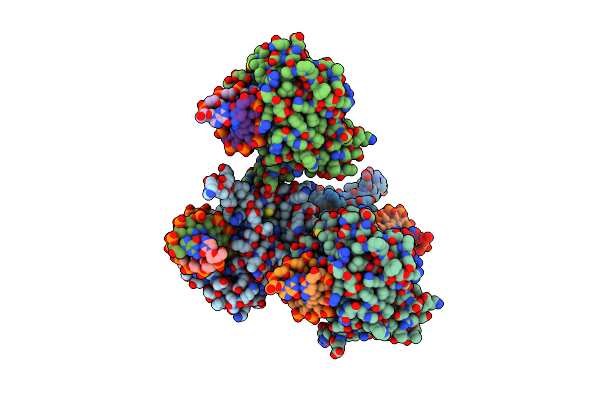
Crystal Structure Of The Tandem Tudor Domains Of The E3 Ubiquitin-Protein Ligase Uhrf1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2008-09-16 Classification: LIGASE Ligands: SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2008-04-29 Classification: LIGASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2007-12-18 Classification: LIGASE Ligands: SO4, EDO, UNL |
 |
Rotation Of The Stalk/Neck And One Head In A New Crystal Structure Of The Kinesin Motor Protein, Ncd
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2003-10-28 Classification: CELL CYCLE Ligands: MG, ADP |
 |
Dimerization Of E. Coli Dna Gyrase B Provides A Structural Mechanism For Activating The Atpase Catalytic Center
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2000-03-31 Classification: ISOMERASE Ligands: SO4, ANP, GOL |

