Search Count: 57
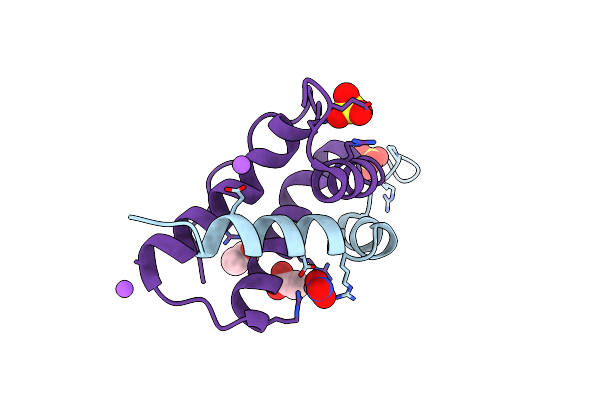 |
Crystal Structure Of Isoform Chitin Binding Protein From Iberis Umbellata L.
Organism: Iberis umbellata
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: PLANT PROTEIN Ligands: SO4, NA, NO3, CL, ACT |
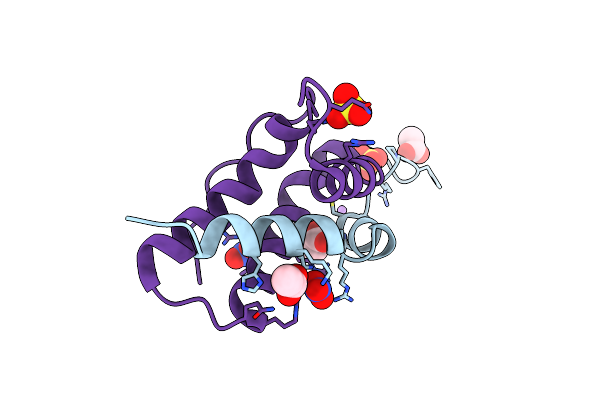 |
High-Resolution Crystal Structure Of An Isoform Of Chitin Binding Protein From Iberis Umbellata L.
Organism: Iberis umbellata
Method: X-RAY DIFFRACTION Resolution:0.91 Å Release Date: 2025-03-19 Classification: PLANT PROTEIN Ligands: SO4, NO3, ACY, LI |
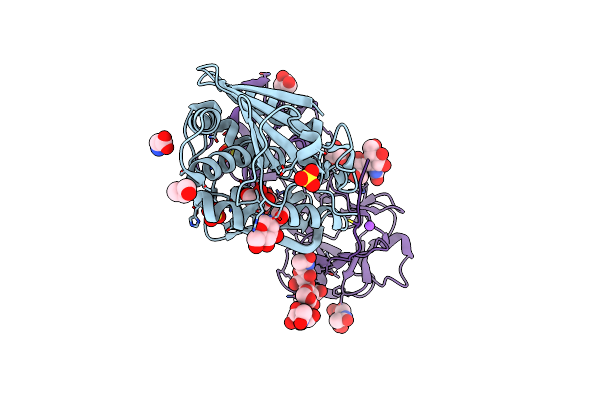 |
Organism: Viscum album
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2025-01-15 Classification: PLANT PROTEIN Ligands: GOL, NAG, SO4, GLY, CL, NA |
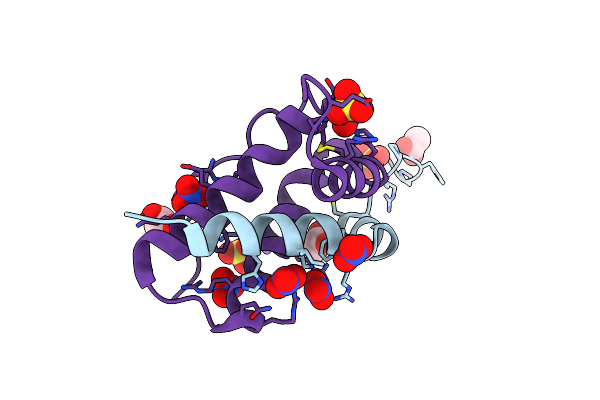 |
Organism: Iberis umbellata
Method: X-RAY DIFFRACTION Release Date: 2024-12-11 Classification: PLANT PROTEIN Ligands: SO4, ACT, NA, NO3, CL |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.83 Å Release Date: 2024-10-02 Classification: TRANSFERASE Ligands: PO4, EDO, CL, MG |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:3.02 Å Release Date: 2024-07-24 Classification: BIOSYNTHETIC PROTEIN Ligands: PO4, GLN, SO4 |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:3.02 Å Release Date: 2024-07-24 Classification: BIOSYNTHETIC PROTEIN Ligands: EDO, PO4, CL, PGE |
 |
Crystal Structure Of Penicillin-Binding Protein 3 (Pbp3) From Staphylococcus Epidermidis
Organism: Staphylococcus epidermidis (strain atcc 35984 / rp62a)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-12-13 Classification: PENICILLIN-BINDING PROTEIN |
 |
Crystal Structure Of Penicillin-Binding Protein 3 (Pbp3) From Staphylococcus Epidermidis In Complex With Vaborbactam
Organism: staphylococcus epidermidis (strain atcc 35984 / rp62a)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-12-13 Classification: PENICILLIN-BINDING PROTEIN Ligands: 4D6 |
 |
Crystal Structure Of Penicillin-Binding Protein 3 (Pbp3) From Staphylococcus Epidermidis In Complex With Cefotaxime
Organism: Staphylococcus epidermidis (strain atcc 35984 / rp62a)
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2023-12-13 Classification: PENICILLIN-BINDING PROTEIN Ligands: CEF |
 |
Crystal Structure Of Sars-Cov-2 Main Protease In Orthorhombic Space Group P212121
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-03-22 Classification: HYDROLASE Ligands: CL, DMS, MLI |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2023-01-25 Classification: VIRAL PROTEIN Ligands: DMS |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2023-01-18 Classification: VIRAL PROTEIN Ligands: CL |
 |
Structure Of Sars-Cov-2 Papain-Like Protease Bound To N-(2-Pyrrolidyl)-3,4,5-Trihydroxybenzoylhydrazone
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2022-03-23 Classification: HYDROLASE Ligands: DZI, GOL, ZN, CL, PO4 |
 |
Structure Of Sars-Cov-2 Papain-Like Protease Bound To N-(3,5-Dimethoxy-4-Hydroxybenzyliden)Thiosemicarbazone
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2022-03-16 Classification: HYDROLASE Ligands: A5I, GOL, ZN, PO4, CL |
 |
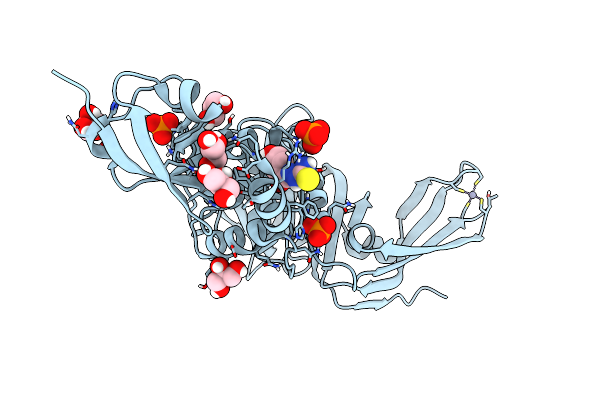
Structure Of Sars-Cov-2 Papain-Like Protease Bound To N-(3,4-Dihydroxybenzylidene)-Thiosemicarbazone
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2022-03-16 Classification: HYDROLASE Ligands: A6Q, GOL, ZN, PO4, CL |
 |
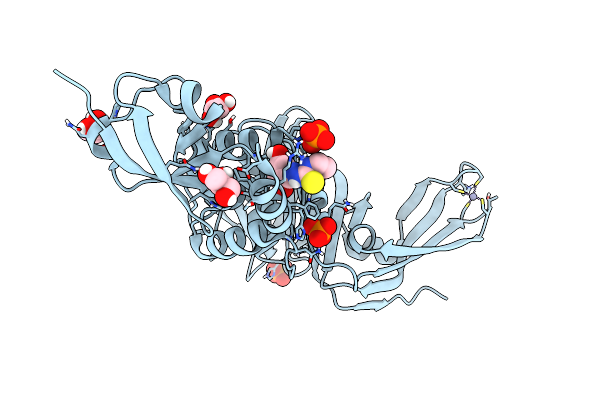
Structure Of Sars-Cov-2 Papain-Like Protease Bound To N-(2,4-Dihydroxybenzylidene)-Thiosemicarbazone
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2022-03-16 Classification: HYDROLASE Ligands: GOL, A4O, ZN, PO4, CL |
 |
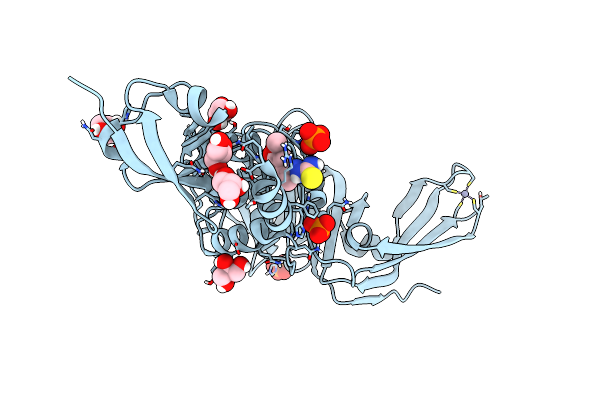
Structure Of Sars-Cov-2 Papain-Like Protease Bound To N-(2,5-Dihydroxybenzylidene)-Thiosemicarbazone
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2022-03-16 Classification: HYDROLASE Ligands: A7L, GOL, ZN, PO4, CL |
 |
Structure Of Sars-Cov-2 Papain-Like Protease Bound To N-(3-Methoxy-4-Hydroxy-Acetophenone)Thiosemicarbazone
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2022-03-16 Classification: HYDROLASE Ligands: A3X, GOL, ZN, CL, PO4 |
 |
Structure Of Sars-Cov-2 Papain-Like Protease Plpro In Complex With 4-(2-Hydroxyethyl)Phenol
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-05-12 Classification: HYDROLASE Ligands: ZN, CL, YRL, PO4, GOL |

