Search Count: 19
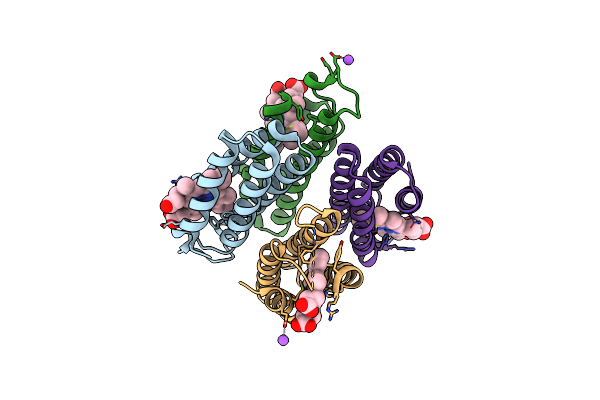 |
Crystal Structure Of An Engineered Metal-Free Ridc1 Variant Containing Five Disulfide Bonds.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-11-09 Classification: ELECTRON TRANSPORT Ligands: HEC, NA |
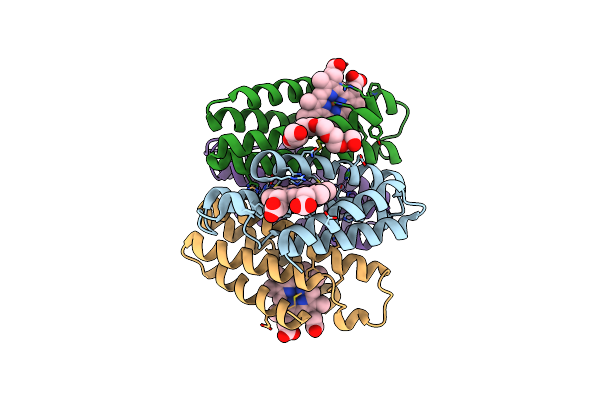 |
Crystal Structure Of The Zn-Ridc1 Complex Bearing Six Interfacial Disulfide Bonds
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-11-09 Classification: METAL BINDING PROTEIN Ligands: HEC, P6G, ZN |
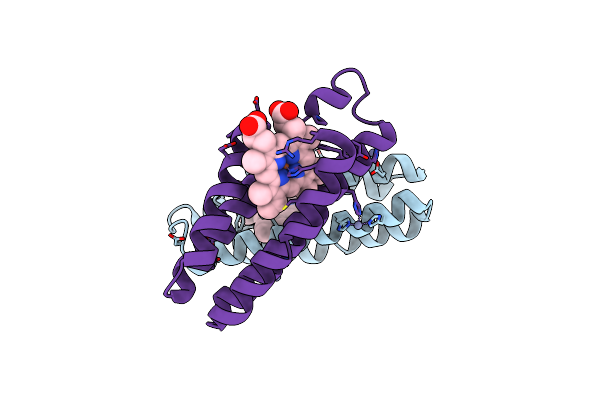 |
Crystal Structure Of An Engineered Protein That Forms Nanotubes With Tunable Diameters
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2016-06-08 Classification: METAL BINDING PROTEIN Ligands: HEM, ZN |
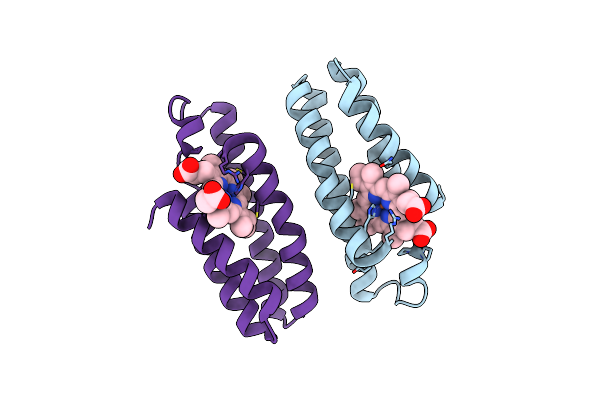 |
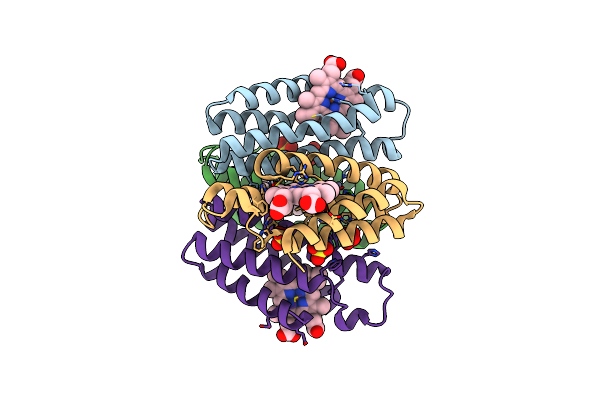
Crystal Structure Of An Engineered Metal-Free Ridc1 Construct With Four Interfacial Disulfide Bonds
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2013-08-21 Classification: ELECTRON TRANSPORT Ligands: HEM |
 |
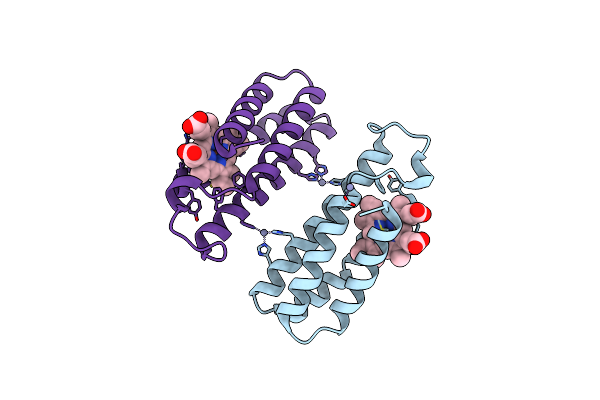
Crystal Structure Of An Engineered Zn-Ridc1 Construct With Four Interfacial Disulfide Bonds
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2013-08-21 Classification: ELECTRON TRANSPORT Ligands: HEM, ZN, SO4 |
 |
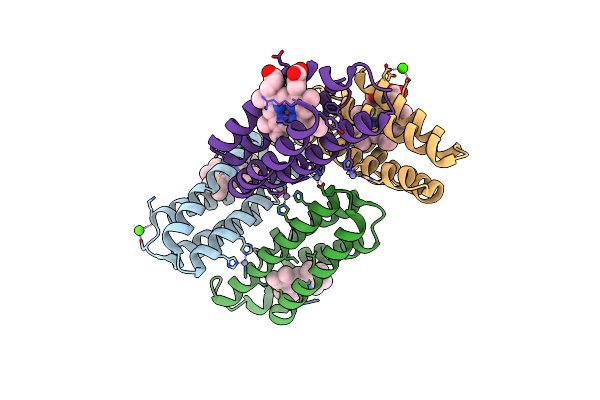
Crystal Structure Of An Engineered Ridc1 Tetramer With Four Interfacial Disulfide Bonds And Four Three-Coordinate Zn(Ii) Sites
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-08-21 Classification: ELECTRON TRANSPORT Ligands: HEM, ZN |
 |
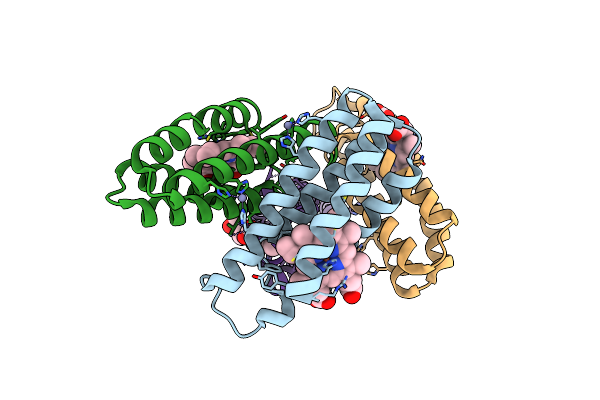
Crystal Structure Of An Engineered Cytochrome Cb562 That Forms 1D, Zn-Mediated Coordination Polymers
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-07-04 Classification: METAL BINDING PROTEIN Ligands: HEM, ZN, CA |
 |
Crystal Structure Of An Engineered Cytochrome Cb562 That Forms 2D, Zn-Mediated Sheets
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-07-04 Classification: METAL BINDING PROTEIN Ligands: HEM, ZN |
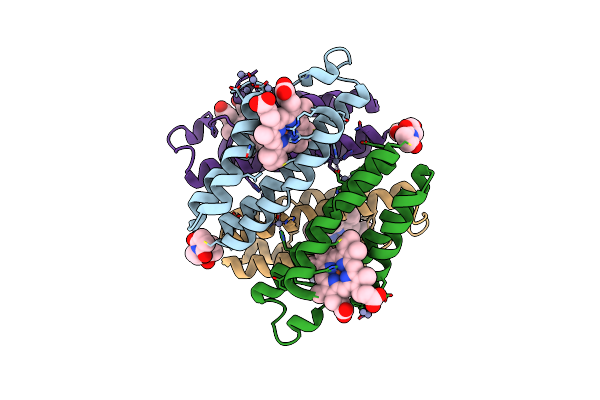 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-06-22 Classification: OXIDOREDUCTASE Ligands: HEM, ZN, ME7 |
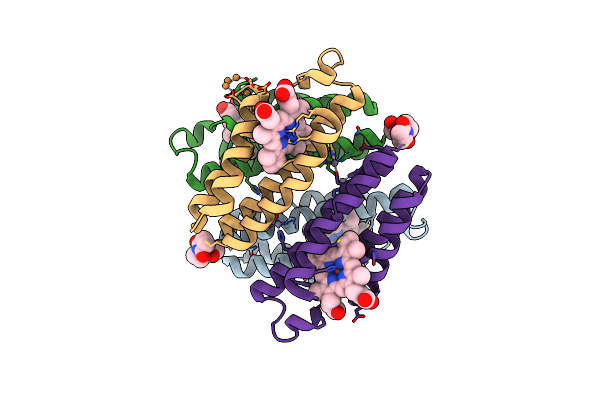 |
Crystal Structure Of The Zn-Ridc1 Complex Stabilized By Bmoe Crosslinks Cocrystallized In The Presence Of Cu(Ii)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2011-06-22 Classification: OXIDOREDUCTASE Ligands: HEM, ME7, ZN, CU |
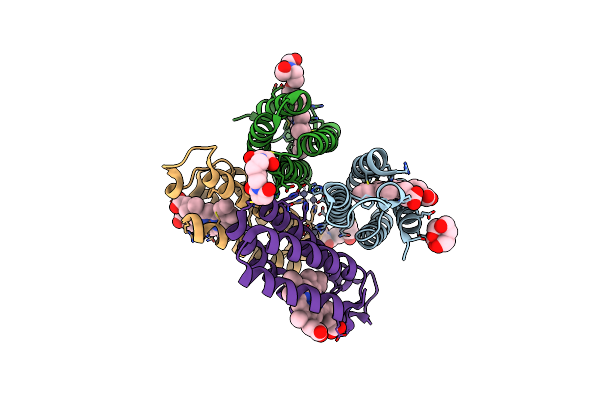 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2011-06-22 Classification: OXIDOREDUCTASE Ligands: HEM, ME9, ZN, BTB |
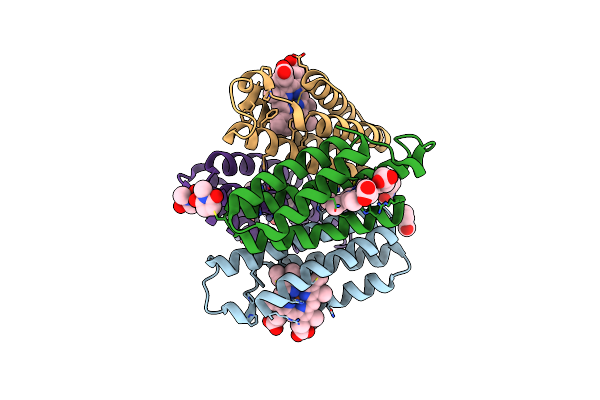 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2011-06-22 Classification: OXIDOREDUCTASE Ligands: ZN, HEM, LCY |
 |
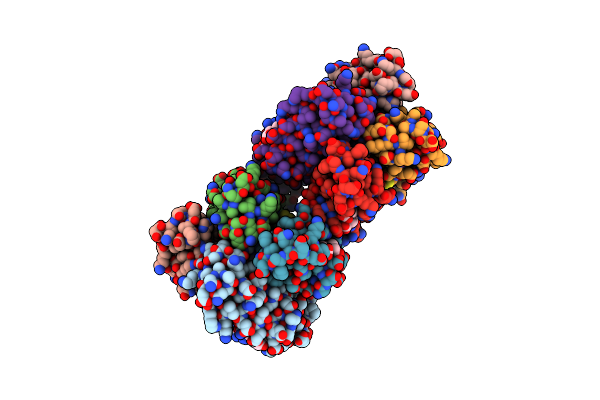
Crystal Structure Of An Engineered Metal-Free Tetrameric Cytochrome Cb562 Complex Templated By Zn-Coordination
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2010-06-16 Classification: ELECTRON TRANSPORT Ligands: HEM |
 |
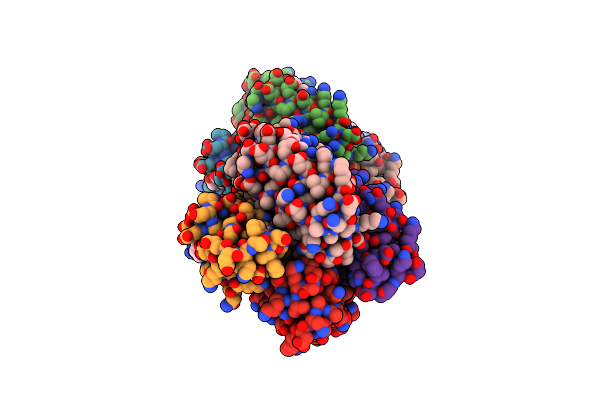
Crystal Structure Of A Tetrameric Zn-Bound Cytochrome Cb562 Complex With Covalently And Non-Covalently Stabilized Interfaces
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2010-06-16 Classification: ELECTRON TRANSPORT Ligands: HEM, ZN |
 |
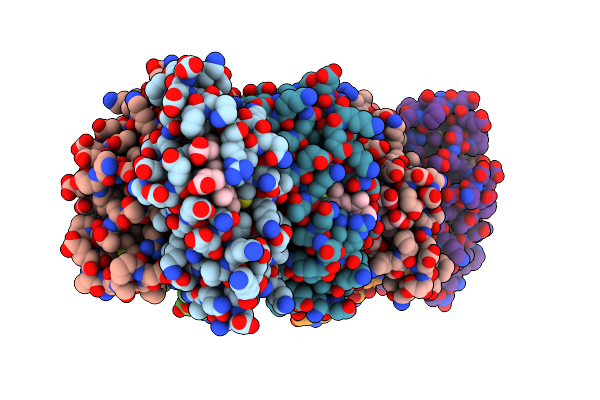
A Tetrameric Zn-Bound Cytochrome Cb562 Complex With Covalently And Non-Covalently Stabilized Interfaces Crystallized In The Presence Of Cu(Ii) And Zn(Ii)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-06-16 Classification: OXIDOREDUCTASE Ligands: HEM, ZN |
 |
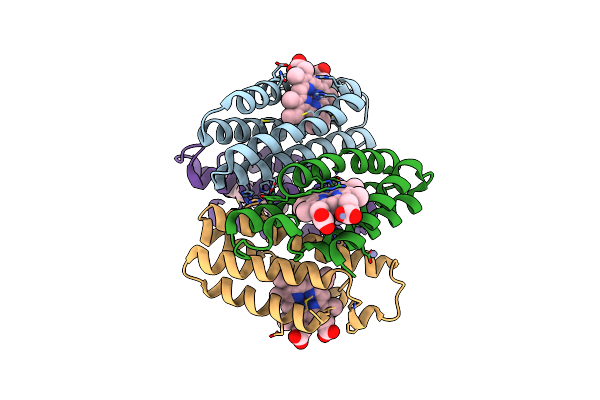
Crystal Structure Of The Zn-Induced Tetramer Of The Engineered Cyt Cb562 Variant Ridc-1
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2010-02-09 Classification: METAL BINDING PROTEIN Ligands: HEM, ZN |
 |
Crystal Structure Of The Zn-Induced Tetramer Of The Engineered Cyt Cb562 Variant Ridc-2
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-02-09 Classification: METAL BINDING PROTEIN Ligands: HEM, ZN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-02-09 Classification: METAL BINDING PROTEIN Ligands: HEM |
 |
Crystal Structure Of The Cu-Induced Dimer Of The Engineered Cyt Cb562 Variant Ridc-1
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-02-09 Classification: METAL BINDING PROTEIN Ligands: HEM, CU, CL |

