Search Count: 76
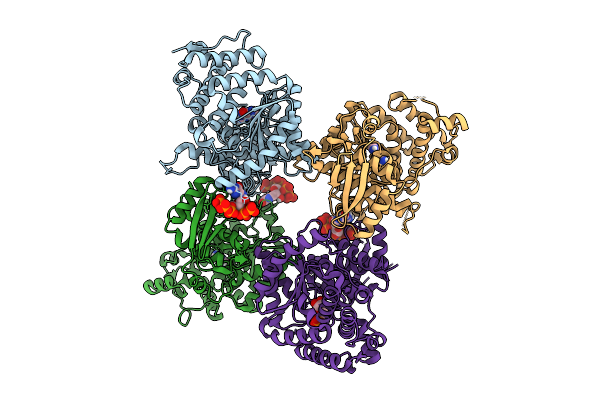 |
Crystal Structure Of The Escherichia Coli Nucleosidase Ppnn (Partial Alarmone Form)
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: 0O2, GUN, HSX |
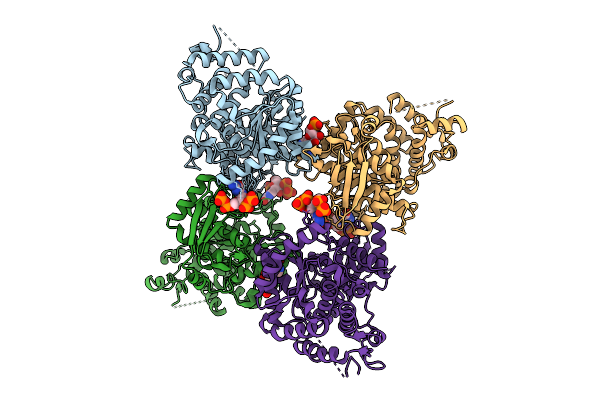 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: G4P, A1IXI |
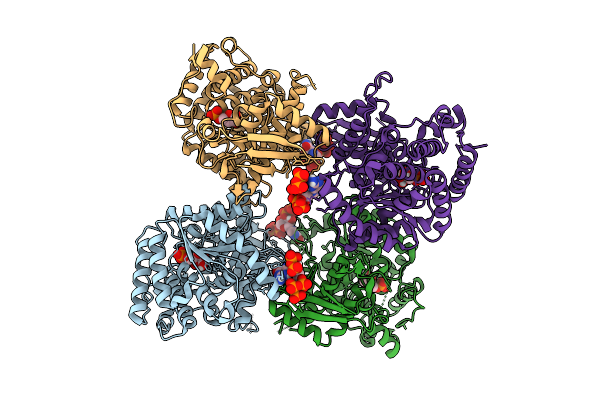 |
Crystal Structure Of The Escherichia Coli Nucleosidase Ppnn (E264A, Pppgpp Form)
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: HSX, 0O2 |
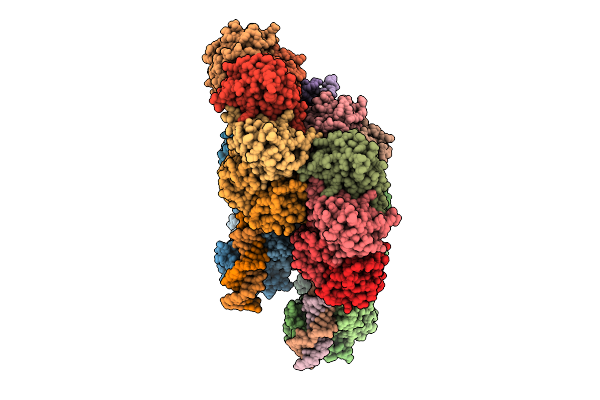 |
Crystal Structure Of The Pseudomonas Putida Xre-Res Toxin-Antitoxin Complex Bound To Promoter Dna
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: GENE REGULATION |
 |
Organism: Rhodonellum psychrophilum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-05-14 Classification: ISOMERASE |
 |
Organism: Rhodococcus sp. jg-3
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2025-05-14 Classification: ISOMERASE Ligands: TRS, NA |
 |
Organism: Escherichia coli kly
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2024-12-18 Classification: TOXIN |
 |
Crystal Structure Of The E. Coli F-Plasmid Vapbc Toxin-Antitoxin Complex (Vapb T3N, A13P, L16R)
Organism: Escherichia coli kly
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-12-18 Classification: TOXIN |
 |
Crystal Structure Of The E. Coli F-Plasmid Vapbc Toxin-Antitoxin Complex (Vapb T3N)
Organism: Escherichia coli kly
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-12-18 Classification: TOXIN |
 |
Crystal Structure Of The E. Coli F-Plasmid Vapbc Toxin-Antitoxin Complex (Vapb V5E)
Organism: Escherichia coli kly
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2024-12-18 Classification: TOXIN |
 |
Se-Met Derivative Structure Of A Small Alarmone Hydrolase (Relh) From Corynebacterium Glutamicum
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-07-13 Classification: HYDROLASE Ligands: MG |
 |
Native Structure Of A Small Alarmone Hydrolase (Relh) From Corynebacterium Glutamicum
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-07-13 Classification: HYDROLASE Ligands: MN, PG4 |
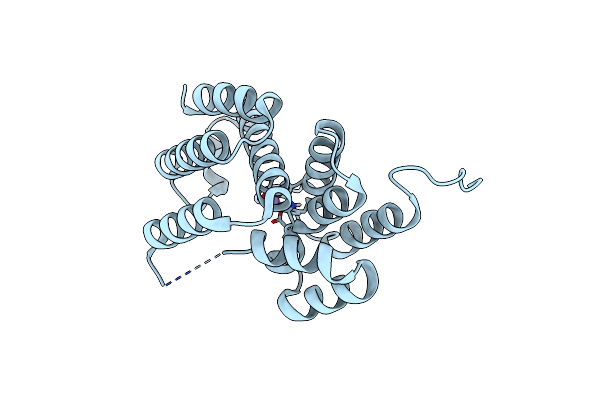 |
Organism: Leptospira levettii
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2022-07-13 Classification: HYDROLASE Ligands: MN |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-06-22 Classification: TRANSFERASE Ligands: ZN, I9X, ADP, PO4, MG, ATP |
 |
E. Coli C-P Lyase Bound To Phnk/Phnl Dual Abc Dimer With Amppnp And Phnk E171Q Mutation
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-06-22 Classification: TRANSFERASE Ligands: ZN, PO4, MG, ANP |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-05-25 Classification: TRANSFERASE Ligands: ZN, I9X |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-05-25 Classification: TRANSFERASE Ligands: ZN, I9X, ATP, MG |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-05-25 Classification: TRANSFERASE Ligands: ZN, PO4 |
 |
Crystal Structure Of The Escherichia Coli Toxin-Antitoxin System Hipbst (Hipt S57A)
Organism: Escherichia coli o127:h6 (strain e2348/69 / epec)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-01-12 Classification: TOXIN |
 |
Crystal Structure Of The Escherichia Coli Toxin-Antitoxin System Hipbst (Hipt S59A)
Organism: Escherichia coli o127:h6 (strain e2348/69 / epec)
Method: X-RAY DIFFRACTION Resolution:3.34 Å Release Date: 2022-01-12 Classification: TOXIN |

